Introduction
Have you ever wondered how tiny organisms like bacteria and viruses can wreak havoc on the human body? Medical microbiology holds the key to unlocking the mysteries of these microscopic villains. Since the dawn of germ theory of disease, medical microbiology has progressed considerably at the end of the last century. It was at this time that premature culture techniques were fruitful in quarantining many pathogens and labeling them with their corresponding causative diseases. Medical microbiology has become an essential component of healthcare and society, thanks to its rapid advancement in response to clinical demands. This field of science investigates the relationship between large and small organisms in both normal and disease conditions, as well as the development of disease and the appropriate treatments that result in complete clinical recovery. A wide range of testing methods are employed to accomplish this task (Savitskaia, 1993). The field of medical microbiology continues to rely heavily on culture-based methods for identifying culture organisms despite substantial developments in analysis methodologies. By identifying the useful microorganisms from those that are pathogenic, medical microbiology contributes to the health of the public and helps to control infectious disease epidemics. Medical microbiology has undergone significant changes in the past few decades, with new methods being developed to make the isolation and detection of various microorganisms more efficient. In medical microbiology laboratories, a range of different microscopy and culture techniques are typically used. One example of a technique that can be used to detect bacteria, fungi, parasites, or viruses in infected cells is indirect fluorescent technique. This technique uses polyclonal antibodies raised in animals like mice or horses, as well as monoclonal antibodies produced using hybridization technology (Sharma et al., 2014). Latex agglutination tests are used to detect particulate antigens, while enzyme immunoassay tests are employed for soluble antigens (House et al., 2005). When it comes to life-threatening infections, it is essential to have quick and accurate diagnostic tests to facilitate prompt antimicrobial therapy (Sharma et al., 2014). In recent times, molecular biology techniques have emerged as a more efficient and rapid way to perform microbiological diagnosis. These methods use nucleic acid probes in conjunction with Polymerase Chain Reaction (PCR) amplification techniques (Ieven et al., 1996). There is a possibility that cloning techniques could be useful in clinical settings, as they are already utilized in different areas of our everyday lives. For instance, during the outbreak of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2), laboratories recognized the need for the swift development of diagnostic tests for emerging diseases that have a significant impact on society. It is vital to quickly transfer such technology to laboratories that perform routine diagnostics. A contemporary issue is that scientific reactions to evolving threats are far more prompt than governmental reactions, and new diagnostic tests for use outside research laboratories often take a long time to be sanctioned (Raoult et al., 2004).
From Past to Present: The Evolution of Molecular Diagnostics
Molecular biology has been a vital tool for the medical laboratory, with its roots dating back to Linus Pauling’s discovery of sickle cell anemia as a “molecular disease” in 1949 (Pauling et al., 1949). However, it was not until recombinant DNA technology in the early days of molecular biology that it became usable in medical diagnostics (Chehab, 1993). Molecular diagnostics grew from basic knowledge on the primary sequence of various genes, with DNA probes incorporating radioactive nucleotides allowing analysis via Southern blotting of genomic regions. This led to the concept and application of restriction fragment length polymorphism (RFLP), which helped track variant alleles in the human genome (Williams, 1989). In 1978, molecular diagnostics techniques were used to make the first prenatal diagnosis of α-thalassemia, and the use of RFLP to characterize sickle cell alleles set the foundation for the characterization of other genetic diseases and infectious diseases using molecular diagnostics platforms (Kan et al., 1978). The mid-1980s brought the development of PCR, which quickly became a staple of laboratory medicine with its ability to exponentially amplify a target sequence and identify known mutations or sequences within hours (Mullis et al., 1986). PCR also established the foundation for many variant detection schemes based on the amplification of DNA. Following the publication of the human genome draft sequence, the challenge to improve existing variant detection technologies to achieve robust, cost-effective, rapid, and high-throughput analysis of genomic variation moved to the forefront of molecular diagnostics. Real-time PCR and its numerous variations, DNA microarray-based genotyping and transcription profiling, microbiome sequencing, proteomics, pharmacogenomics, nutrigenomics, forensic medicine, and CRISPR/Cas9 genome editing all represent important and critical advances in the field (Hendrix and Rohde, 2021). Despite the explosion of diverse variant detection assays, DNA sequencing remains the gold standard for pathogen identification and surveillance, especially with breakthroughs in next-generation sequencing (NGS) technology (Hendrix and Rohde, 2021). However, the costs of initial investment and difficulties in standardization and interpretation of ambiguous results continue to limit the use of NGS in clinical laboratories. Physicians and other healthcare professionals are now working with molecular diagnostics professionals to understand the basis of infectious disease pathology and when to use molecular diagnostics like NGS. One example is the use of 16S in-house assay sequencing to identify bacterial pathogens directly from tissue specimens when culture results are negative, but there is evidence of histopathologic pathogen damage (Hendrix and Rohde, 2021). In addition to PCR and sequencing, molecular cloning also played a crucial role in the development of molecular diagnostics techniques. By enabling the replication of specific DNA sequences, molecular cloning made it possible to generate large amounts of identical DNA fragments for further analysis. This technique is based on the use of restriction enzymes to cut DNA at specific recognition sites and then ligating these fragments into a plasmid vector for amplification and manipulation. The ability to produce recombinant proteins using molecular cloning revolutionized the field of medical research and led to the production of many important biological therapeutics, including insulin, growth hormone, and clotting factors. Molecular cloning also allowed for the creation of genetic probes that could be used to detect specific DNA sequences, such as those associated with infectious agents. This enabled the development of highly sensitive and specific diagnostic tests, such as the PCR-based tests that are widely used today for the detection of viruses like HIV and hepatitis C. Molecular cloning techniques have also facilitated the identification of new disease-causing genes and the development of gene therapy approaches for genetic diseases.
An Overview of Molecular Cloning
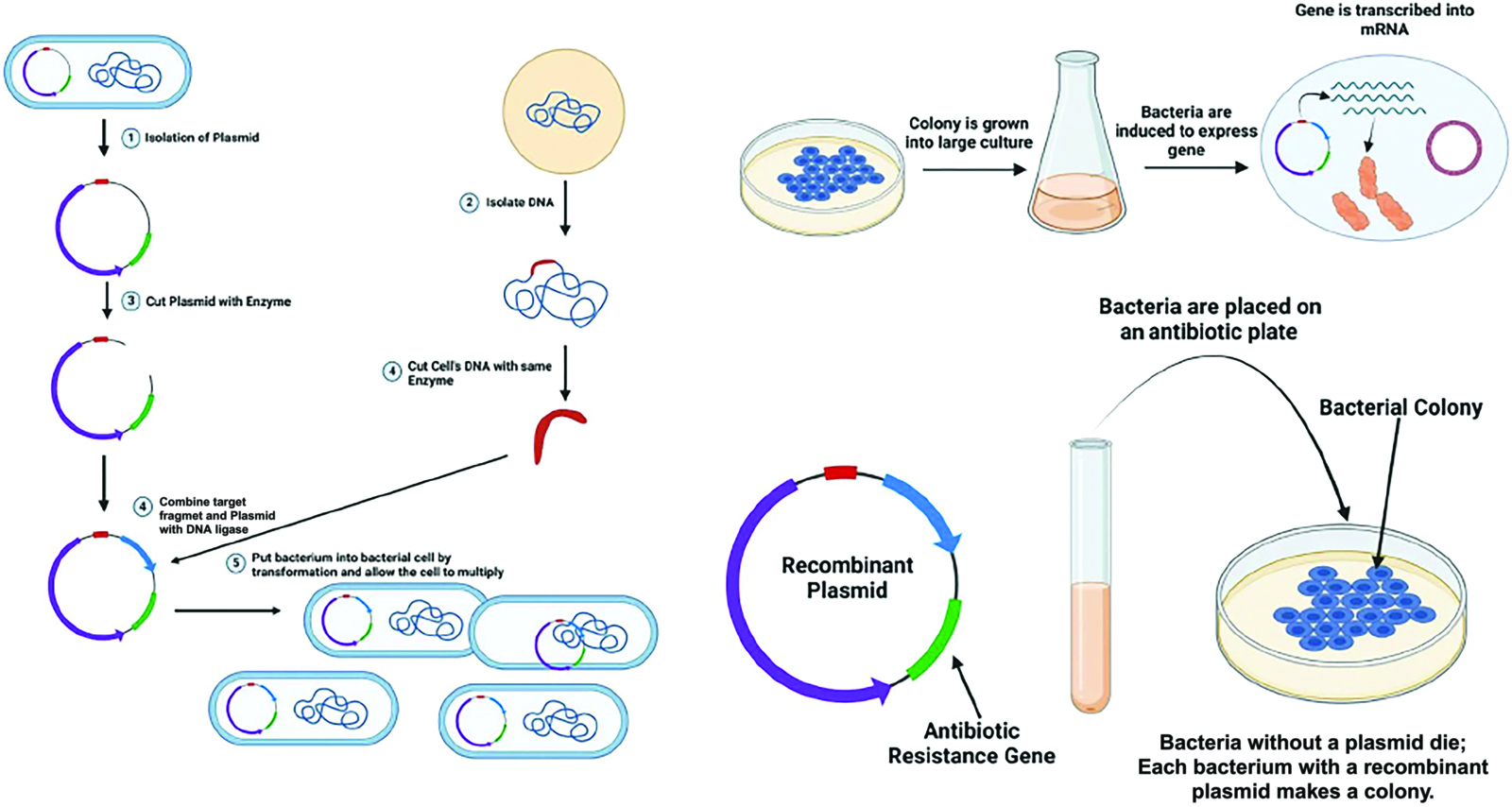
In the annals of scientific progress, the field of molecular cloning stands as a testament to human ingenuity and perseverance. Throughout history, researchers have endeavored to unlock the secrets of DNA, seeking ways to manipulate and understand its intricate structure. It is within this context that the technique of molecular cloning emerged, revolutionizing the study of genetics and paving the way for a multitude of biological and technological applications. Molecular cloning (a molecular diagnostics technique) refers to a technique that involves isolating a specific DNA sequence and then amplifying short regions of it in vitro. The roots of molecular cloning can be traced back to the mid-20th century, when scientists began unraveling the mysteries of DNA. It was during this time that the concept of isolating specific DNA sequences and amplifying them in vitro took shape. One commonly employed approach involved the use of restriction enzymes, which acted as molecular scissors, cutting DNA at specific sites. By digesting existing DNA fragments or targeting them through PCR, researchers were able to generate short inserts of DNA, typically around 100 base pairs in length (Juliane and Lessard, 2013). These resulting short inserts can also be created as complementary single-stranded fragments that are then annealed to form a double-stranded fragment (Juliane and Lessard, 2013). Once the DNA of interest has been obtained, it can be inserted into a vector plasmid, which is a circular double-stranded DNA. These vectors are smaller versions of naturally occurring plasmids and contain features such as replication origins, drug resistance genes, and unique restriction sites that enable the insertion of DNA fragments (Juliane and Lessard, 2013). The multiple cloning sites of these vectors usually contain different restriction sites, making it easier to select the appropriate enzymes for a variety of inserts. Molecular cloning was typically used to amplify DNA fragments containing genes, but it can also be used to amplify any DNA sequence, including promoters, non-coding sequences, chemically synthesized oligonucleotides, or fragments of randomly generated DNA (Sharma et al., 2014).
There was a wide range of biological applications and technological applications that make use of this method, including the production of recombinant antigens, cytokines, and proteins (Nguyen et al., 2004). If DNA sequences were to be amplified and cloned in vitro and in vivo, they must be linked to primary sequence elements that are capable of directing their own replication and propagation in the desired target host in conjunction with the linked sequence (Lu et al., 2008). Thus, the inclusion of a host-specific origin of replication and a selectable marker became essential sequence elements. In addition, when selecting a cloning vector, researchers had to consider a number of other characteristics, including the ability to express proteins, tag them, and generate single-stranded RNA and DNA (Sharma et al., 2014). As the field progressed, researchers sought to refine and expedite the cloning process. Recombinase-based cloning emerged as a one-step reaction, allowing for high-throughput cloning by inserting a specific DNA fragment into a specific region of target DNA through the interchange of relevant DNA fragments (Copeland et al., 2001). This streamlined approach facilitated the cloning of any DNA fragment, representing a significant leap forward compared to the classical restriction- and ligation-based approach, which involved fragmenting DNA with restriction endonucleases, ligating it to a vector, transfecting it into host cells, and subsequently screening and selecting the desired clones (Sharma et al., 2014). Cloning procedures generally followed these classical steps; however, a number of unconventional routes can be chosen depending on the specific solicitation.
Cloning procedures had traditionally adhered to the classical steps outlined previously, but the field of molecular cloning continued to evolve, offering researchers a growing repertoire of unconventional routes to choose from, based on the specific goals and requirements of their studies. Some advancements had further expanded the possibilities and enhanced the efficiency of molecular cloning techniques. One notable development in molecular cloning was the emergence of advanced DNA assembly methods that enable the construction of large DNA constructs with precise control over their sequence composition. For instance, techniques such as Gibson assembly, Golden Gate assembly, and ligase cycling reaction (LCR) had revolutionized the process by allowing seamless assembly of multiple DNA fragments in a single reaction (Engler et al., 2008; Gibson et al., 2009; Li and Elledge, 2007). These methods bypassed the labor-intensive steps of traditional restriction digestion and ligation, streamlining the process and reducing the occurrence of unwanted mutations. Gibson assembly was a method that allowed the seamless assembly of multiple DNA fragments without the need for restriction enzymes or DNA ligases. It operated on the principle of in vitro homologous recombination and utilized three key enzymatic activities: exonuclease, polymerase, and DNA annealing. In this technique, the DNA fragments to be assembled were designed with overlapping regions called “homology arms.” These homology arms enabled the fragments to anneal to one another in the presence of the exonuclease and polymerase enzymes, which trimmed back the ends of the fragments and filled in the gaps, respectively. The resulting annealed fragments were then extended and ligated, producing a seamless composite DNA construct (Gibson et al., 2009). Gibson assembly offered several advantages, including its simplicity, efficiency, and the ability to seamlessly assemble multiple fragments with high fidelity, minimizing the introduction of errors. Golden Gate assembly was another powerful technique used for the modular assembly of DNA fragments. It relied on the activity of type IIS restriction enzymes, such as BsaI, which cut DNA sequences outside their recognition sites. The DNA fragments to be assembled were designed with specific recognition sequences for the type IIS enzyme at their ends, along with overlapping regions. During the assembly process, the type IIS restriction enzyme cuts the DNA fragments at the specific recognition sites, generating cohesive ends. The cohesive ends from different fragments were then ligated together, creating a composite DNA construct (Engler et al., 2008). Golden Gate assembly offered advantages such as modularity, versatility, and compatibility with high-throughput applications. It allowed the construction of complex DNA constructs by assembling multiple fragments in a single reaction, enabling researchers to efficiently generate diverse genetic constructs with precise control. LCR was a technique that combined the principles of PCR and DNA ligation to facilitate the assembly of DNA fragments. In LCR, short DNA oligonucleotides, called splints, were designed to anneal to specific regions on the DNA fragments to be ligated. These splints hybridized to their complementary sequences, bringing the DNA fragments in close proximity for ligation. The ligation step was mediated by a DNA ligase enzyme, which catalyzed the formation of phosphodiester bonds between the adjacent DNA fragments. The ligated DNA fragments were then amplified by PCR using primers that hybridized to the ends of the assembled construct (Li and Elledge, 2007). These advanced molecular cloning techniques had transformed the field by offering streamlined processes for the assembly of multiple DNA fragments. They eliminated the need for laborious traditional methods, such as restriction digestion and ligation, and provided advantages such as increased efficiency, reduced errors, and the ability to construct complex DNA constructs with precision.
Another significant advancement was the utilization of site-specific recombinases, such as the Cre-Lox system and the Flp-FRT system, to enable precise DNA rearrangements. These recombinases catalyzed the exchange, inversion, or deletion of DNA segments at specific target sites, facilitating the generation of complex DNA constructs and genetic modifications (Sauer and Henderson, 1988; Buchholz et al., 1996). The Cre-Lox system, derived from the bacteriophage P1, consisted of two key components: the Cre recombinase enzyme and the LoxP sites. In the Cre-Lox system, the Cre recombinase recognized and bind to LoxP (locus of crossover [X] P1) sites present in the DNA. The LoxP site consisted of two 13-base-pair inverted repeats flanking a central 8-base-pair spacer region. The Cre recombinase binds to the LoxP site and catalyzes a strand cleavage and exchange reaction. This reaction involves the cleavage of the DNA strands at the specific LoxP sites, followed by the exchange or rearrangement of the cleaved DNA segments. The result was the excision, inversion, or rearrangement of DNA segments depending on the orientation and arrangement of the LoxP sites (Sauer and Henderson, 1988). The Cre-Lox system had been widely used for conditional gene knockout, gene expression control, and the generation of tissue-specific or inducible genetic modifications. This system offered precise control over DNA modifications, as the recombination events occurred exclusively at the LoxP sites and did not affect surrounding genomic regions. Similarly, the Flp-FRT system, derived from the yeast Saccharomyces cerevisiae, operated through the interaction between the Flp recombinase enzyme and the FRT (Flippase Recognition Target) sites. FRT sites are DNA sequences that serve as recognition sites for the Flp recombinase. The FRT site consisted of two 13-base-pair inverted repeats separated by a central 8-base-pair spacer region. The Flp recombinase recognized the FRT site and catalyzed a similar strand cleavage and exchange reaction as the Cre-Lox system. By cleaving the DNA strands at the FRT sites and facilitating the exchange or rearrangement of the cleaved segments, the Flp-FRT system enabled the excision, inversion, or rearrangement of DNA segments, depending on the arrangement and orientation of the FRT sites (Buchholz et al., 1996).The Flp-FRT system offered high precision and versatility in genetic manipulations, which allowed researchers to precisely control the outcome of the recombination events. The benefits of these site-specific recombinase systems are manifold. First, they offered precise control over DNA modifications, allowing researchers to manipulate specific regions within a genome without affecting surrounding genetic elements. This targeted approach minimized unwanted off-target effects and preserved the integrity of the genome. Second, these systems provided flexibility and versatility in creating complex genetic modifications, such as gene knockouts, gene insertions, and conditional gene expression systems. The ability to precisely control DNA rearrangements at specific target sites opened up new avenues for functional studies and understanding gene regulation. Moreover, the reversible nature of these recombinase systems allowed for the creation of conditional genetic modifications, where DNA segments can be switched between different configurations, providing temporal and spatial control over gene expression.
In recent years, CRISPR-Cas9 technology has revolutionized molecular cloning and genetic engineering. Originally developed as a precise genome editing tool, CRISPR-Cas9 has been adapted for DNA cloning applications, allowing for efficient and targeted insertion of DNA fragments into specific genomic loci (Kleinstiver et al., 2014; Nishimasu et al., 2014). The CRISPR-Cas9 system has simplified and accelerated the process of creating transgenic organisms, generating knock-in or knock-out models, and facilitating the study of gene function (Doudna and Charpentier, 2014). The working principle of the CRISPR-Cas9 system involves two main components: the Cas9 enzyme and a guide RNA (gRNA). The Cas9 enzyme acts as a molecular scissor and can be programmed to target specific DNA sequences with the help of the gRNA. The gRNA is designed to recognize a complementary DNA sequence adjacent to the target site, guiding the Cas9 enzyme to that specific location. Once the Cas9 enzyme binds to the target DNA sequence, it generates a double-strand break (DSB) at that site. This DSB triggers the cellular DNA repair machinery, which can be harnessed to introduce desired DNA fragments into the genome (Doudna and Charpentier, 2014). CRISPR-Cas9 technology offers several significant benefits in molecular cloning and genetic engineering. First and foremost, it provides unparalleled precision in DNA targeting. The gRNA can be easily customized to recognize any desired DNA sequence, allowing researchers to selectively edit or insert DNA fragments at specific genomic loci. This level of precision enables the creation of highly specific genetic modifications, facilitating the study of gene function and the elucidation of molecular mechanisms. Another advantage of CRISPR-Cas9 technology is its efficiency and versatility. The Cas9 enzyme, guided by the gRNA, efficiently generates DSBs at the target site, triggering the cellular repair mechanisms. Researchers can exploit these mechanisms to introduce exogenous DNA fragments into the genome. This enables the precise insertion of DNA fragments, such as genes, regulatory elements, or reporter constructs, at desired genomic locations. In addition, the system can be used for gene knockouts by introducing DSBs that lead to gene disruptions or deletions. CRISPR-Cas9 technology has significantly streamlined the process of creating transgenic organisms and generating genetically modified cell lines, providing researchers with a powerful tool for studying gene functions and disease mechanisms. The advancements in synthetic biology have opened new avenues for molecular cloning. DNA synthesis technologies have improved significantly, enabling the de novo assembly of entire genes, pathways, or even genomes (Kosuri and Church, 2014). This has paved the way for the creation of synthetic DNA constructs with tailored functions, such as metabolic pathways for bioproduction, novel enzymes, or even synthetic organisms with redesigned genomes (Gibson et al., 2010; Hutchison et al., 2016). These recent developments in molecular cloning techniques have not only increased efficiency but also expanded the possibilities for genetic manipulation, DNA engineering, and biotechnological applications. By embracing these cutting-edge approaches, researchers can push the boundaries of what is achievable in the field of molecular cloning.
The Role of Molecular Cloning in Polymicrobial Infections
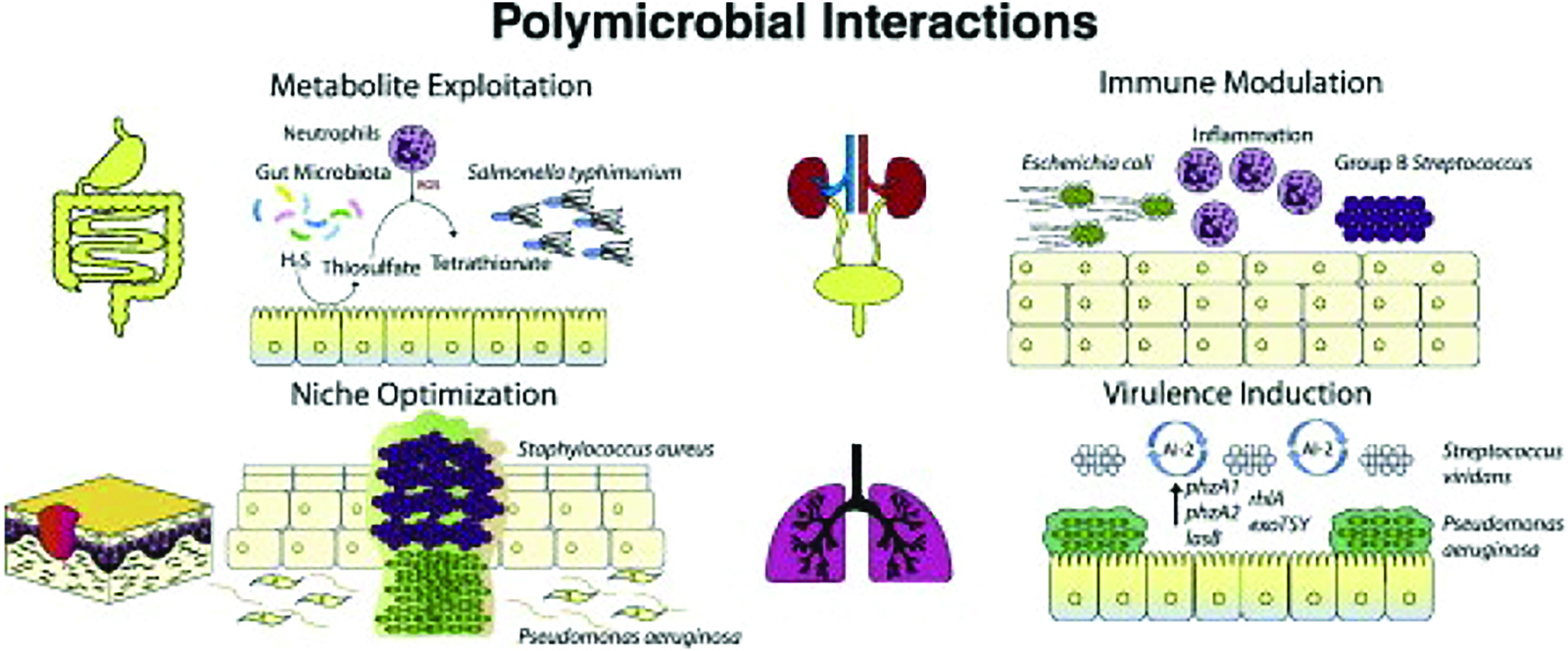
Polymicrobial infections, where multiple microorganisms are involved in an infection, are a common occurrence in clinical settings. Molecular cloning had become an important tool in identifying diseases caused by multiple microorganisms and understanding the exchanges that occur within microbial communities. These diseases, as described by Kim et al. (2005), can be acute or chronic and are caused by combinations of viruses, bacteria, fungi, and parasites (Sharma et al., 2014). When a single pathogenic microorganism creates a niche, other pathogenic microorganisms can inhabit it, leading to colonization or the emergence of disease by two or more non-pathogenic microorganisms (Kim et al., 2005). Culture-based routine diagnostic testing is one treatment strategy, but it has limitations since it may not isolate all significant microbe species present in a sample. New bacterial community profiling techniques have revealed a greater diversity of microbes in infections caused by these bacteria than previously thought (Sharma et al., 2014). It is the result of these findings that polymicrobial infections are increasingly being perceived as complex communities of interacting organisms, whose pathogenicity is determined by dynamic processes (Rogers et al., 2010). One of the primary applications of molecular cloning in polymicrobial infections is the identification and characterization of individual pathogens. Traditional microbiological methods involve culturing the microorganisms from clinical samples, which can be time-consuming and may not always yield accurate results. In contrast, molecular cloning techniques such as PCR and NGS can identify and differentiate multiple pathogens in a single sample rapidly and accurately. These techniques can identify pathogens that are difficult or impossible to culture, making them a valuable tool in the diagnosis of polymicrobial infections. However, even these techniques have certain drawbacks (Datta, 2023).
In the past few decades, advances in cloning and NGS technology had provided us with opportunities for gaining such insights about viable microbial cells (Wheat, 2010). An approach based on sequence homology allowed identification of bacteria in polymicrobial infections by cloning and sequencing the 16S ribosomal gene (Amann et al., 1995). With this method, it was possible to identify bacteria that died in the course of transportation or due to antibiotic treatment and to discover bacteria with specific growth requirements (Kommedal et al., 2009). However, it may not be possible to detect rare members of a community with divergent target sequences with rRNA gene-based cloning and characterization (Petrosino et al., 2009). Some of the limitations for it are due to primer bias and low sampling depth, which could be solved by 454 sequencing, pyrosequencing, or whole genome shotgun sequencing (Petrosino et al., 2009). Hence, molecular cloning techniques, such as Loop-mediated isothermal amplification (LAMP), Multi-Plex PCR-Based Reverse Line Blot Hybridization (mPCR-RLB), Target Enriched-Multiplex PCR (Tem-PCR), Gene chip technology, and Multiplex Ligation-Dependent Probe Amplification (MLPA), are utilized, which play a crucial role in identifying the genes and pathways involved in polymicrobial infections (Datta, 2023). These techniques enable researchers to gain a comprehensive understanding of the mechanisms underlying the interactions between pathogens, which is essential for developing novel treatments that target these interactions. LAMP is a powerful nucleic acid amplification technique that allows for the detection and quantification of specific DNA sequences at a constant temperature. LAMP amplifies DNA by exploiting the properties of Bst polymerase and the unique structure of the target DNA. It involves the design of specific primers for different regions of the target gene, resulting in the formation of a loop structure. The amplification reaction produces a characteristic ladder-like structure (amplicon), which can be visualized using fluorescence or turbidity changes (Datta, 2023). LAMP is particularly useful for the prompt recognition of pathogenic microorganisms in laboratories with limited resources and experimental conditions. mPCR-RLB combines the sensitivity and the specificity of PCR with the high-throughput capabilities of reverse line blot hybridization. It involves the use of multiple primer sets that specifically bind to conserved regions of the microbial genome (Datta, 2023). These primers are used in a multiplex PCR reaction to amplify multiple target DNA sequences simultaneously. The amplified DNA is then hybridized to a reverse line blot membrane containing probes for different microbes. The presence or absence of hybridization signals allows for the identification of specific microbial types in the sample. Tem-PCR combines the specificity of PCR with target enrichment to detect and identify microbes involved in polymicrobial infections (Datta, 2023). The target enrichment step involves the selective amplification of specific regions of interest from the sample DNA using specific probes. This is followed by a multiplex PCR reaction, where multiple sets of primers amplify different targets in the same reaction. Tem-PCR allows for the simultaneous detection of multiple targets with high sensitivity and specificity. This technique is particularly useful for the detection of low-abundance targets in clinical samples. Gene chip technology, also known as DNA microarray, is a powerful tool for the detection and characterization of microbes. It allows for the simultaneous detection and analysis of multiple microbial genes, providing a comprehensive view of viral load and genetic diversity (Datta, 2023). Gene chips consist of a glass slide or silicon wafer coated with thousands of DNA probes complementary to specific regions of the microbial genome to be tested. When a sample of genomic DNA or cDNA is labeled and hybridized to the chip, the binding of target sequences to the complementary probes generates fluorescent complexes. The resulting signals provide information on the relative expression levels of the genes in the sample, enabling the identification of differentially expressed genes and grouping of samples based on their gene expression profiles. MLPA is a technique used to detect and quantify specific sequences in a sample. It combines the specificity of PCR with ligation and probe amplification. MLPA involves the ligation of two probes, one specific for the target sequence and the other for a control sequence, followed by PCR amplification using universal primers (Datta, 2023). The resulting pool of amplified fragments, proportional to the amount of DNA in the sample, can be quantified and analyzed to determine relative copy numbers of target sequences.
Molecular cloning also plays a crucial role in understanding the interactions between multiple pathogens in polymicrobial infections. For example, studies have shown that certain bacterial species can facilitate the growth of other pathogens, which can lead to more severe infections (Doron and Gorbach, 2008). Molecular cloning techniques can help identify the genes and pathways involved in these interactions, allowing researchers to better understand the mechanisms behind polymicrobial infections. This understanding can lead to the development of new treatments that target the interactions between pathogens rather than just targeting individual pathogens. Once the key genes and pathways responsible for pathogen interactions have been elucidated through molecular cloning techniques, researchers can delve deeper into understanding the underlying mechanisms. Armed with this knowledge, they can develop innovative treatments specifically designed to disrupt the cooperative or synergistic relationships between the pathogens involved. Several novel treatment strategies have emerged from this approach, each targeting different aspects of pathogen interactions. One such strategy involves interfering with quorum sensing, a cell-to-cell communication system employed by many bacteria to coordinate their activities (Rutherford and Bassler, 2012). Quorum sensing is a cell-to-cell communication system utilized by many bacteria, enabling them to synchronize their gene expression and coordinate their activities as a group (Rutherford and Bassler, 2012). This coordinated behavior often leads to the production of virulence factors, biofilm formation, or the regulation of key processes required for the survival and colonization within the host. Researchers have been able to identify the genes and molecules involved in quorum sensing through molecular cloning techniques, and by understanding the specific components of the quorum sensing system, they can develop therapeutic agents that interfere with this communication process and disrupt the signaling and coordination among pathogens (Vadakkan et al., 2018). There are different strategies employed to interfere with quorum sensing. One approach involves blocking or inhibiting the production or activity of quorum sensing signaling molecules, such as autoinducers (Vadakkan, 2020). These signaling molecules are produced by bacteria and accumulate in the environment as the population density increases and they act as chemical signals, and when their concentration reaches a threshold, they trigger specific gene expression programs, coordinating behaviors among the bacterial community (Vadakkan, 2020). Therapeutic agents targeting quorum sensing can be designed to mimic or block the action of these signaling molecules (Milly and Tal‐Gan, 2023). For example, small molecules can be developed that either competitively bind to the receptor sites of bacteria, preventing the binding of natural signaling molecules, or mimic the signaling molecules themselves, leading to false or ineffective coordination among the pathogens (Milly and Tal‐Gan, 2023). Another strategy involves inhibiting the production or activity of the enzymes responsible for synthesizing the signaling molecules, which results in the disruption in communication and coordination among pathogens (Paluch et al., 2020). This interference can be achieved through the development of enzymatic inhibitors or by targeting the genes encoding the enzymes involved in the synthesis process (Paluch et al., 2020). Such an approach holds promising potential in combating pathogenic infections by impeding their ability to communicate and coordinate their detrimental actions.
The Role of Molecular Cloning in Antimicrobial Peptides
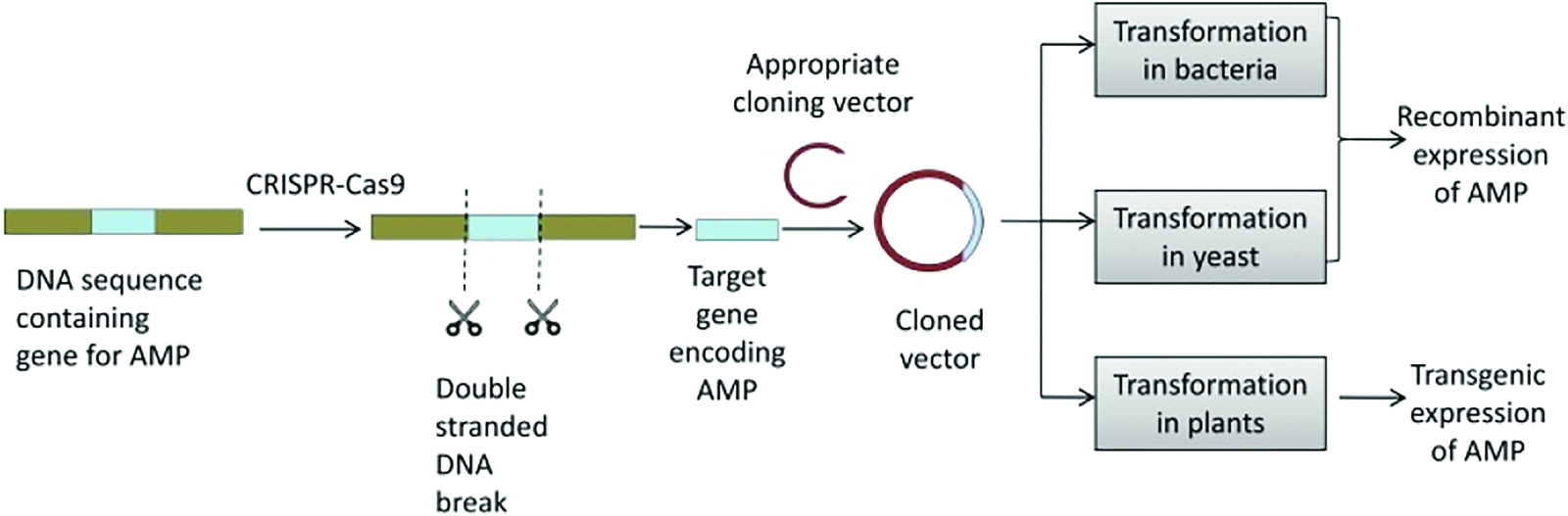
In order to combat the rapidly increasing antimicrobial resistance, more effective antimicrobial peptides are being developed. These antimicrobial peptides will kill target cells promptly and be effective against antibiotic-resistant and clinically pertinent pathogens. Advances in molecular genetics of antibiotic biosynthesis offer new opportunities to improve antibiotic production. The DNA of antibiotic makers is enriched with genes coding for antibiotic biosynthesis enzymes. There have been two types of vectors developed to clone antibiotic genes: low-copy and high-copy plasmids (Fakruddin et al., 2013). Many manufacturers are actively registering molecular clones of antibiotic synthesis genes. By manipulating the cloned genes encoding the enzymes involved in the biosynthetic pathway, new antibiotics can be produced as structural deviations of existing ones (Muller et al., 2007). These structural variants have diverse spectra and potency of activity against innumerable bacteria (Sharma et al., 2014). Moreover, the production of highly purified antimicrobial peptides at competitive costs is one of the biggest challenges facing the development of antimicrobial peptides.
Chemical peptide synthesis can be used to yield either native or modified cationic peptides, but it is more expensive than isolating peptides from natural sources. In vivo synthesis in host cells using recombinant technology is a more effectual and cost-effective method of synthesis (Muller et al., 2007). Recombinant expression of antimicrobial peptides is a promising area of research despite the challenges posed by their toxicity to bacteria and susceptibility to degradation by proteases (Sharma et al., 2014). This technology is widely recognized as an effective method for enhancing protein, peptide, and enzyme production. It offers advantages such as reduced time, well- established protocols, cost-effectiveness, and scalability (Sinha and Shukla, 2019). Bacteria and yeast are the most commonly used host systems for expressing recombinant products, including antimicrobial peptides (AMPs) (Gupta and Shukla, 2017). Escherichia coli, particularly the strain E. coli BL21 (DE3), is a popular choice due to its fast growth rate, high yields, established expression protocols, and commercial availability of expression vectors. Other bacterial systems like Bacillus subtilis have also been used although to a lesser extent. Among yeast, Pichia pastoris has been employed as a potential host. Scientists have developed strategies to address these issues, such as fusing peptide genes with larger proteins and then using enzymes or chemicals to cleave them and release active peptides (Kuhnel et al., 2003). Promoter probe vectors are also being employed to clone DNA sequences with transcriptional control signals. One interesting approach involves neutralizing the positive charge of the peptide to enhance its activity (Ali and Murrell, 2009). Another promising application of molecular cloning is the amplification of genes involved in biosynthesis pathways. By increasing the production of limiting enzymes, researchers have shown that it is possible to boost antibiotic production. This represents a significant step forward in the fight against antibiotic-resistant bacteria, which pose a major threat to public health. With the help of molecular cloning techniques, scientists are now better equipped to identify and develop novel antimicrobial agents that can be used to combat infectious diseases. Recombinant expression of AMPs is a popular method to produce large quantities of these peptides for further study or therapeutic use. However, the toxicity of some AMPs against bacteria and their susceptibility to proteolytic degradation can present challenges in the expression process (Nizet, 2006). To overcome these issues, researchers have used molecular cloning techniques to fuse the AMP genes with larger proteins. The fusion proteins can then be cleaved by enzymes or chemicals to release active AMPs (Ingham and Moore, 2007). To mitigate the toxicity of AMPs to the host strain, fusion proteins consisting of the target AMP and a carrier protein are often used. Carrier proteins possess anionic properties that neutralize the cationic charge of AMPs, reducing their toxicity and increasing solubility (Li, 2009). Common fusion partners include thioredoxin, Small Ubiquitin-like Modifier (SUMO), Glutathione S-transferase (GST), Biotin Carboxyl Carrier Protein (BCCP), and Green Fluorescent Protein (GFP). Cleaving the carrier protein from the target AMP typically requires the use of chemicals or enzymes. Chemical cleavage, although less specific, is considered more efficient than enzymatic cleavage, but it can introduce modifications in the side chain (Sinha and Shukla, 2019). Cleavage processes often leave one or two non-native residues at the N-terminus of the cleaved AMP (Sinha and Shukla, 2019). Affinity tags are sometimes conjugated to fusion partners to facilitate easy purification using affinity chromatography methods (Meiyalaghan et al., 2014). Examples include His6-thioredoxin tagged GSL1 fusion protein, which can be purified by affinity chromatography, and fusion partners like glutathione S-transferase, which have inherent affinity properties and eliminate the need for additional tags (Schäfer et al., 2015). Self-cleaving tags with inducible proteolytic activity have also been used to simplify the separation of fusion tags. Thioredoxin and SUMO are the preferred fusion partners for the expression of recombinant AMPs (Xia et al., 2013). For instance, cecropin XJ, an insect AMP, was highly expressed in E. coli as a fusion peptide along with thioredoxin (Sinha and Shukla, 2019). Similarly, LsGRP1C protein production was assisted by a yeast SUMO tag in the E. coli host system, resulting in a high yield of the soluble SUMO-LsGRP1C fusion protein (Lin et al., 2017). BCCP has been used as a fusion protein for AMP expression, and B. subtilis has been employed for the recombinant expression of cathelicidin-BF from snake venom (Luan et al., 2014, Orrapin and Intorasoot, 2014). Another method for cloning AMPs is through the use of promoter probe vectors. These vectors contain transcriptional control signals that can be used to clone DNA sequences with transcriptional control signals by neutralizing the positive charge of the peptide (Muriana and Klaenhammer, 1991). This allows for more efficient expression of AMPs in various hosts. Furthermore, molecular cloning has also been used to amplify genes coding for limiting enzymes in biosynthesis pathways, which has been shown to increase the production of antimicrobial compounds (Adrio and Demain, 2010). This approach has led to the discovery and production of new antimicrobial compounds with potent activity against a number of microorganisms. Yeasts, particularly the methylotrophic yeast Pichia pastoris, have become increasingly important in genetic engineering and recombinant protein production due to their ease of genetic manipulation, ability to perform complex post-translational modifications, and rapid growth in cost-effective media (Kim et al., 2015). P. pastoris is a popular host for heterologous expression of recombinant AMPs (Ahmad et al., 2014). It offers advantages over E. coli, including the presence of a methanol-induced alcohol oxidase promoter, absence of endotoxins, correct protein folding capability, and suitability for large-scale production (Ahmad et al., 2014). In one study, the NZ17074 gene was synthesized and fused with SUMO3 in P. pastoris X-33, and the carrier protein was subsequently cleaved using formic acid (Wang et al., 2014). AMPs from plants, fruits, and chicken have also been expressed in P. pastoris but without the use of fusion proteins (Meng et al., 2017). The emergence of advanced gene editing tools, including Zinc-Finger Nucleases (ZFNs), Transcription Activator-Like Effector Nucleases (TALENs), and Clustered Regularly Interspaced Short Palindromic Repeat-CRISPR-associated protein (CRISPR-Cas), has opened up new possibilities in the field of gene editing (Dangi et al., 2018). These tools make it easier to manipulate the genomes of expression hosts, enabling targeted modifications of specific genes to achieve desired outcomes. By harnessing gene editing technology, it is now possible to revolutionize the production of AMPs, particularly in response to the increasing demand for industrially and therapeutically valuable AMPs (Sinha and Shukla, 2019).
The Role of Molecular Cloning in Recombinant Cytokines
There are several fundamental homeostatic mechanisms that are modulated by cytokines (Huang et al., 2005). They include fever, acute phase infections, wound healing, inflammation, immune responses at the cellular levels, and tumor deterioration. Recombinant DNA technology has allowed us to clone the genes that encode these proteins, making it possible to use unrestrained measures of these cytokines to treat disease. There can be changes in amino acid sequence, absence of glycosylation (E. coli), and changes in glycosylation pattern (yeast, mammals, and insects) (Bandaranayake et al., 2011). Proteins expressed in the mature form in different host cells can also differ in their specific activities for several reasons (Sharma et al., 2014). Studies show that cytokines expressed in different host cells can have different pharmacokinetics, biologic properties, and immunogenicity due to physiochemical differences (Descotes, 2009). To explore the structure-function relationship of cytokines, expression vectors can be used to hypothesize recombinant forms. In addition to cytokines engineered for improved clinical efficiency or novel specificities, heterologous expression systems have also been used to create streamlined cytokines (Bermúdez-Humarán et al., 2011). Nowadays, recombinant cytokines are available as therapeutic agents. There has been evidence that GM-GSF or G-CSF can reduce the period and risk of infectious complications associated with chemotherapy-induced neutropenia (Sharma et al., 2014). Molecular cloning techniques are utilized to produce recombinant cytokines and enable their mass production. One of the most commonly produced cytokines through molecular cloning is interferon- alpha (IFN-α), which has potent antiviral and antiproliferative properties (Kumar et al., 2018). Recombinant IFN-α has been used in the treatment of hepatitis B and C infections, as well as certain types of cancer, including melanoma and leukemia. Molecular cloning is also used to produce other cytokines, such as interleukins and tumor necrosis factor (TNF). Recombinant TNF has been used in the treatment of bladder cancer, while interleukins have been used to enhance the immune response against cancer cells (Muller et al., 2003). In addition, the use of cytokines produced through molecular cloning has been explored in the treatment of autoimmune diseases, such as rheumatoid arthritis and multiple sclerosis. Moreover, the investigation into the therapeutic potential of molecularly cloned cytokines has extended to encompass the management of autoimmune disorders, including rheumatoid arthritis and multiple sclerosis. These chronic conditions, such as rheumatoid arthritis, are characterized by persistent joint damage caused by an overactive immune response targeting the synovial membrane, cartilage, and bone (Zhang, 2021). While the precise cause of rheumatoid arthritis remains elusive, significant progress has been made in understanding its complex pathogenesis involving various cell types and signaling pathways (Verhoef et al., 2019).
Autoimmune processes and cytokines have emerged as key players in the initiation and perpetuation of rheumatoid arthritis. Notably, the extensively studied tumor necrosis factor (TNF), interleukin-6 (IL-6), and interleukin-1 (IL-1) have been implicated in the disease progression (Upchurch and Kay, 2012). Consequently, therapeutic strategies have focused on combating inflammation, with non-steroidal anti-inflammatory drugs and glucocorticoids serving as primary symptomatic management options (Zhang, 2021). In recent years, disease-modifying antirheumatic drugs (DMARDs) have revolutionized rheumatoid arthritis treatment by specifically targeting pro-inflammatory cytokines and their respective receptors (O’Dell, 2004). These biological DMARDs include TNF-α inhibitors (such as infliximab, etanercept, adalimumab, golimumab, and certolizumab pegol), IL-1 inhibitors, monoclonal antibodies against the IL-6 receptor (tocilizumab), T cell signaling inhibitors (abatacept), and monoclonal antibodies targeting CD20 (rituximab) (Klarenbeek et al., 2011). When combined with conventional synthetic DMARDs, early initiation of these treatments has shown promising results, leading to improved clinical outcomes and reduced joint damage (Zhang, 2021).
Along with using recombinant immunomodulators to enhance the host’s defense mechanism, attempts have been made to repackage defective immune genes (Miyake et al., 2001). The gene therapy of adenosine deaminase deficiency (ADA) deficiency has successfully treated patients with a defective T cell immune response (Aiuti and Roncarolo, 2009). It is possible to transfer a cloned ADA gene into lymphocytes of a patient using a retroviral vector; enzymatic and immune functions are thus reinstated (Aiuti and Roncarolo, 2009). Many ongoing research projects focus on gene therapy’s use in HIV infection and oncogenic virus-associated cancer, even though there is no clear evidence of its impact on infectious disease. Through HBV- or HHV-8-encoded surface receptors, the thymidine kinase gene has been inserted into the cancer cell in HBV-associated hepatocellular carcinomas and HHV68-associated Kaposi’s sarcomas (Sharma et al., 2014). The patient is given ganciclovir when the thymidine kinase gene is integrated into their cancer cells, where the drug amasses and becomes toxic (Kieback et al., 2008). Future developments in the creation of cytokine receptor agonists show promise in a number of ways. One avenue of exploration involves using de novo protein design in addition to the existing combinatorial ligand engineering strategies. De novo design involves a computational approach to design analogues of interleukin-2 (IL-2) and interleukin-15 (IL-15) that can signal independently of IL-2Rα/IL-15Rα (Saxton et al., 2023). These synthetic analogues, known as “neoleukins,” have unique structural features compared to natural cytokines. They exhibit enhanced stability and improved effectiveness in mouse tumor models. Currently, neoleukins are undergoing phase I clinical trials for multiple cancer types, either alone or in combination with immune checkpoint blockade therapies. Advancements in artificial intelligence and machine learning-based protein structure prediction are expected to facilitate the development of even more sophisticated de novo designed cytokines (Saxton et al., 2023). These cytokines may have entirely distinct structural topologies that can modulate receptor geometry and composition in novel ways, potentially revolutionizing our understanding of cytokine signaling. While de novo cytokines offer advantages such as enhanced stability and potential ease of manufacturing, they may carry a risk of increased immunogenicity due to their non-human amino acid sequences (Saxton et al., 2023). However, ongoing efforts focus on minimizing immunogenicity by reducing the number of mutations and utilizing databases to assess potential neo-epitopes. Improvements in the pharmacokinetic and pharmacodynamic properties of cytokines are also crucial for their clinical success and strategies like half-life extension through Fc fusions or PEGylation, as well as local cytokine production via engineered T cells, aim to enhance cytokine safety and efficacy (Saxton et al., 2023). However, these modifications can influence cytokine signaling activity, necessitating a thorough understanding of their effects. Experimental models for screening and testing novel cytokine activities include in vitro cell culture and mouse models, but these have limitations in predicting human efficacy. Human patient-derived organoid cultures with diverse immune cell populations may provide valuable insights into the effectiveness of cytokine-based drugs in humans (Saxton et al., 2023). Immunogenicity is another significant consideration for protein therapeutics, including engineered cytokines. Non-human proteins, like de novo designed cytokines, may carry a higher risk of eliciting immune responses due to their divergent amino acid sequences, so efforts to minimize immunogenicity, especially for mutant cytokines, include reducing the number of mutations and monitoring for cross-neutralizing antibodies (Saxton et al., 2023).
The Role of Molecular Cloning in Gene Therapy
An individual can be treated with gene therapy by introducing a regular gene into their genome in order to mend the mutation that is the origin of genetic disease. In addition to possibly repairing the mutation, insertion of a regular gene into another functional gene may result in a new mutation if the normal gene integrates into another functional gene’s chromosomal site (Khan et al., 2016). Transformed cells may proliferate if normal genes replace mutant genes, leading to the restoration of the non-disease phenotype of the entire body. To date, human gene therapy has only been tested on somatic cells to treat cancer and severe immunodeficiency syndromes (Khan et al., 2016). It is possible for gene therapy to inverse the signs of disease in somatic cells, but the adjustment does not pass on to future generations. By placing corrected cells inside the germ line (e.g., cells of the ovary or testis), gene therapy will ensure that the next generation of cells undergoes meiosis and contributes to standard gametic development (Khan et al., 2016). In the field of health services, gene therapy is a progressive technique that has therapeutic prospect. The first successful report in gene therapy for the cure of genetic diseases gave physicians a promising approach to treating the lethal genetic disorders (Cavazzana-Calvo et al., 2000). Treatment for the primary immunodeficiency adenosine deaminase-deficiency (ADA-SCID) shows good results with this approach. An improved gene transfer protocol and myeloablative conditioning regime, however, were later used to achieve fruitful results by aiming the hematopoietic stem cells (HSCs) (Aiuti et al., 2002).
The expression of specific genes by lentiviral vectors can correct X-linked disorders and adrenoleukodystrophy (X-ALD) (Cartier et al., 2009). Based on HIV-1 genes, X-ALD protein expression indicates gene-correction of true HSCs. As part of the treatment of metastatic melanoma through immunotherapy, lentiviral vectors were used for the first time in the cure of genetic human diseases (Montini et al., 2012). By expanding the field of health sciences through immunotherapy, new opportunities were unlocked for treating serious diseases that cause death (Morgan et al., 2006). In two patients, continuous levels of T cells engineered to recognize tumors in the blood after infusion resulted in the recession of metastatic melanoma lesions up to a year after infusion. The bioengineered T cells were later studied for the treatment of chronic lymphocytic leukemia and metastatic synovial cell carcinoma where autologous T cells were innately altered to express Chimeric Antigen Receptors (CARs) that specifically bind to B cell antigen CD19 (Robbins et al., 2011). It has shown remarkable results for incorrigible autosomal recessive dystrophies, such as congenital blindness and Leber congenital amaurosis (LCA), in which gene transfer to a small number of cells at anatomically discrete sites has the potential to confer therapeutic benefit. Through gene therapy, a variety of cancers have been treated, including lung, gastrointestinal, hematological, gynecological, skin, urological, and neurological tumors (Khan et al., 2016). In order to treat diverse types of cancer, tumor suppressor genes have been inserted into immunotherapy, oncoturlytic virotherapy, and gene-directed enzyme prodrugs. In some cancer treatment strategies, p53 gene transfer is combined with chemotherapy or radiotherapy to increase the effectiveness of the tumor suppressor gene. An effective new anticancer agent (Ad5/35-EGFP) is being developed from fiber chimeric adenovirus vectors for the improved cure of hepatocellular carcinoma (Khan et al., 2016). In hepatocellular carcinoma (HCC), these vectors were established to improve transduction and produce more virus progeny as a consequence of proper assaying. As a result of complex transgenic expression, in vitro HCC cells were found to possess enhanced antitumor activity while normal cells remained cytotoxic-free. As a result of the use of this technology, tumor growth was also repressed (Zhang et al., 2011). Recently, cancer gene therapy has gained more advanced technology and expanded its effectiveness (Lam et al., 2013). A mutation of the ABCA1 gene in high-density lipoproteins can cause the cells to discriminate into macrophages. Knockouts of this gene in embryonic stem cells augment the capability of these cells to differentiate into macrophages and precisely aim abnormal cells. A study of these allele replacements will provide insights into the regulatory variants that alter macrophage transcription and stability of mRNA (Smith, 2016).
Yan et al. (2020) have spearheaded a groundbreaking advancement in molecular biology by introducing a cutting-edge technique called “Nimble Cloning.” This pioneering method revolutionizes standardized molecular cloning, a fundamental technology in the field. By harnessing the combined power of the restriction enzyme SfiI and the T5 exonuclease, Nimble Cloning enables simultaneous vector linearization and generation of 3’-overhangs. Notably, this novel cloning system accommodates both PCR products and plasmids as inputs for the cloning reaction, rendering it highly efficient and adaptable to gene expression in both prokaryotic and eukaryotic systems. What sets Nimble Cloning apart is its remarkable versatility and simplicity. It empowers researchers to reuse DNA fragments or plasmid entry clones, facilitating efficient and streamlined workflows. This innovative method proves to be equally adept at cloning single or multiple fragments, as well as facilitating multi-site cloning. Consequently, the possibilities for modular assembly of DNA constructs are greatly expanded. In their groundbreaking research, Yan et al. (2020) introduced Nimble Cloning, a novel technique inspired by Gibson assembly, for the seamless assembly of DNA fragments. By leveraging the power of enzyme-catalyzed reactions, they demonstrated that Nimble Cloning surpasses traditional Gibson assembly in terms of cloning efficiency. Excitingly, this method eliminates the need for Taq DNA ligase, streamlining the process without compromising effectiveness. Through meticulous experimentation with single and multiple DNA fragments, Yan et al. successfully validated the superior performance of Nimble Cloning, consistently achieving over 99% positive clones. To further emphasize its versatility, they employed unique adapters in standardized cloning reactions, confirming that gene expression remained unaffected in both prokaryotic and plant systems. These findings not only establish Nimble Cloning as an efficient and reliable method for genetic engineering but also pave the way for future advancements in DNA fragment assembly.
In recent years, the CRISPR-Cas9 system has revolutionized the field of gene therapy. This revolutionary gene editing tool enables precise and efficient modification of specific DNA sequences within the genome. Molecular cloning plays a critical role in the construction of CRISPR-Cas9 vectors, which are used to deliver the Cas9 nuclease and guide RNA sequences into target cells. The ability to edit genes with unprecedented precision has opened up new possibilities for treating genetic diseases by correcting or modifying disease-causing mutations. Advancements in DNA synthesis technologies have facilitated the design and construction of synthetic genes and gene circuits for gene therapy applications. Molecular cloning techniques allow the assembly of these synthetic genes into expression vectors, enabling the production of therapeutic proteins or the regulation of gene expression in a controlled manner. In terms of gene delivery, tissue-specific targeting has become an area of active research, and by engineering viral and non-viral vectors to possess tissue-specific promoters or ligands, researchers aim to enhance the specificity and efficiency of gene delivery to target tissues or cells. This targeted approach minimizes off-target effects and improves the overall safety and efficacy of gene therapy. Innovations in genome engineering techniques, such as base editing and prime editing, have expanded the possibilities of gene therapy. These advanced tools enable precise modification of individual bases within the genome, offering the potential to correct disease-causing mutations without the need for introducing foreign DNA.
The Role of Molecular Cloning in Epidemiology
The rise of multidrug-resilient pathogens necessitates early molecular epidemiology outlining, both for comprehensive public-health reconnaissance and for timely treatment of infested patients. Due to the inoculum size and culturing conditions’ inconsistency, conservative tests of this type require extended culturing times (48–72 h) (Yang and Rothman, 2004). As genetic mechanisms of drug resistance are explained, nucleic-acid-based assays are being developed to report these inadequacies (Yang and Rothman, 2004). Three examples of how molecular epidemiology can be applied clinically are provided next. Although the absence of a resistance gene does establish a lack of resistance through that particular genetic mechanism, the presence of a resistance gene does not ineludibly infer its expression and conferment of phenotypic resistance: for example, the mecA gene is responsible for methicillin resistance (Yang and Rothman, 2004). With its high sensitivity and specificity, the mecA-PCR has become the most consistent method for detecting methicillin-resistant staphylococcus aureus (MRSA) due to its high detection sensitivity (Tenover et al., 1999) The detection of rifampicin resistance in M tuberculosis can also be performed using PCR-based resistance testing. RNA polymerase resistance in M tuberculosis is well characterized and is conferred by mutations within the DNA-directed RNA polymerase subunit beta (rpoB) gene, which result in amino acid substitutions in the rpoB subunit (Telenti et at.,1997). The Line Probe assay (LiPA; Inno-Genetics) targets the mutation-prone rpoB gene segment (De Beenhouwer et al., 1995). A dramatic waning in detection time is provided to clinicians with this method, which is acute for treatment decisions, with an association of over 90% with typical resistance-detection methods (Marttila et al., 1999). There are many mutations and genetic loci involved in other M tuberculosis drug resistances, which makes genotyping more challenging (Yang and Rothman, 2004). Technical innovations like multiplex PCR or DNA microarray allow concurrent extension and analysis of multiple target sequences and will likely be able to overcome this future challenge (Elnifro et al., 2000) A PCR followed by nucleotide sequencing method is currently the most habitually used process to identify drug-resistant mutations in HIV-infected patients with ever-increasing indication supporting their prediction value (Demeter and Haubrich, 2001). Even though genotypic tests are more multifaceted than typical antimicrobial susceptibility tests, they provide perception into resistance development by perceiving mutations at concentrations too low to affect phenotypic assay susceptibility (Yang and Rothman, 2004). Also, they have the benefit of detecting mutations that do not cause drug resistance but do designate selective drug pressure, which could influence treatment decisions for individual patients.
A recent study by Zhang et al. (2021) showed that the pathogen Carbapenem-Resistant Acinetobacter baumannii (CRAB) poses a significant threat to both health and economy. It haunts hospitals, using them as breeding grounds for its insidious transmission. The vulnerable victims of CRAB’s infections are often those who find themselves confined within hospital walls, with open wounds and lengthy stays. The elderly, whose natural defenses have weakened, are especially susceptible. Strikingly, most patients in the study suffered from multiple diseases simultaneously, with pulmonary infections, such as pneumonia and respiratory failure, being the most common. This aligns with the well-known association between CRAB and pneumonia and bloodstream infections. The key to CRAB’s resistance lies in its ability to produce carbapenemases, such as blaOXA-23 and blaOXA66, which render it impervious to carbapenem antibiotics (Zhang et al., 2021). This study by Zhang et al. (2021) confirmed that all strains harbored these carbapenem resistance genes. The mechanisms behind CRAB’s increased antibiotic resistance are multifaceted, involving mobile genetic elements, chromosomal β-lactamases like blaADC, and the presence of efflux pumps (Zhang et al., 2021). The study also highlighted the presence of various genetic structures, including ISs and AbaR-type genomic resistance islands, which facilitate the spread of antibiotic resistance determinants among pathogens, compromising treatment efficacy. But CRAB is not merely resistant; it is also armed with virulence factors that allow it to thrive in the inhospitable environment of a hospital. The study also revealed that all CRAB strains possessed a repertoire of virulence genes, such as bap, csuABCD, pgaABCD, bfmRS, entE, ompA, and plcD. These genes contribute to the formation and maintenance of biofilms, which are strongly associated with MDR strains and clinical infections. In addition, CRAB isolates carried genes involved in the production and uptake of the acinetobactin siderophore, further enhancing their infectious prowess (Zhang et al., 2021). When it comes to the origins of CRAB outbreaks, molecular cloning holds the key to unraveling the mystery. Previous reports have linked clonal outbreaks to international clone lineages, with European clones I, II, and III being the most prominent. In the study conducted by Zhang et al. (2021), all CRAB isolates belonged to ST2, which is associated with outbreaks and harbors the blaOXA-23 gene. Interestingly, certain clone types, such as ST2, ST25, and ST78, exhibited enhanced biofilm formation, likely contributing to their success in colonizing the clinical environment (Zhang et al., 2021). Through the lens of core genome phylogenetic analysis, it became clear that specific CRAB strains dominated the scene. Cluster 1 and cluster 2 emerged as the primary players, with clone 1 and clone 2 persisting throughout the study period (Zhang et al., 2021). However, changes in clone groups were observed over the years, indicating the dynamic nature of CRAB populations. These clones, particularly clone 1, underwent population expansion and exhibited distinct resistance, virulence, and insertion sequence patterns compared to clone 2 (Zhang et al., 2021). These findings emphasize the genetic diversity within CRAB outbreaks and the importance of monitoring and controlling the spread of specific clones.
The Role of Molecular Cloning in Bioterrorism
Biological warfare, in its essence, is the strategic utilization of living organisms or their by-products to cause harm or destruction. It involves the deliberate release or deployment of pathogens, toxins, or biological agents with the intent to incapacitate, debilitate, or even kill living organisms, including humans. This form of warfare relies on exploiting the natural abilities of organisms to produce toxins or propagate diseases, harnessing their destructive potential as a means of achieving military or political objectives. Biological warfare encompasses a range of tactics, from the use of infectious diseases as weapons to the manipulation of toxic substances derived from a number of organisms. Its history intertwines with humanity’s understanding of the natural world and our capacity to wield its forces for destructive purposes. An anthrax outbreak that occurred has drawn much attention to the growing threat of bioterrorism. Responsibility of the clinicians will be crucial in instigating applicable response actions if they can recognize and diagnose real or suspected bioterrorism events quickly and accurately (Pavlin et al., 2002). It is difficult to discriminate between a bioterrorism victim’s symptoms and symptoms of an ordinarily encountered disease process, as was the case at the time of the 2001 anthrax episode (Pavlin et al., 2002). In suspected clinical outbreaks, conventional culture-based assays cannot detect bioterrorism agents due to the formerly designated restrictions. As a result of the prolonged incubation required by conventional microbiological methods, the laboratory is exposed to increased biohazard risks because of the redundant proliferation of bioterrorism pathogens (Yang and Rothman, 2004). Many bioterrorism agents have, of late, been studied using PCR-based assays, including Variola major, Bacillus anthracis, Yersinia pestis, and Francisella tularensis (Espy et al., 2002). Bioterrorism agent PCR diagnostics was used for both screening preclinical victims for early prophylactic treatment and diagnosing symptomatic individuals (Yang and Rothman, 2004). Despite the similarity between most bioterrorism-induced illnesses and natural outbreaks, it was possible that the contributory agents of bioterrorism may have been genetically engineered to be more virulent, impervious to antibiotics or vaccines, or to produce phenotypic characteristics resembling multiple infections via the insertion of recombinant genes (Alibek, 1999). Since DNA-based methodologies are more easily adjustable and proficient in uncovering more comprehensive information embedded within genetic sequences, they are likely to be more valuable than conventional detection methods in such cases (Yang and Rothman, 2004).
The potential role of genetic engineering in enhancing the lethality of infectious agents is a topic often discussed in relation to biological warfare. While there is some validity to this claim, it is crucial to consider the following perspective: Imagine a scenario where a bioterrorist endeavors to genetically modify a harmless laboratory bacterium, such as E. coli. The aim would be to render the bacterium “invisible” to the human immune system upon entry into the body (Clark and Pazdernik, 2016). In addition, the bacteria could be engineered to release toxins, thwarting immune cells and introducing genes to impede the vital iron supply from blood cells. Lastly, the bacterium could be modified to possess high levels of infectivity. Such a biological agent would undoubtedly pose a formidable threat. Astonishingly, this bacterium already exists in nature—it is known as Yersinia pestis, the notorious causative agent of bubonic plague (Clark and Pazdernik, 2016). Endemic in different parts of the world, including China, India, Madagascar, and the United States, this pathogen demonstrates how nature has provided an exceptionally dangerous biological weapon. Consequently, the notion of enhancing infectious diseases through genetic engineering appears to be a minor concern. Although information regarding the Soviet germ warfare facility’s modification of the smallpox virus and the creation of artificial mutants and hybrids remains largely undisclosed, recent experiments involving mousepox (Ectromelia virus) have yielded alarming outcomes. Mousepox, a virus primarily infecting mice, exhibits varying degrees of virulence depending on the mouse strain. Genetically resistant mice rely on cell-mediated immunity rather than antibodies to combat the virus, with natural killer (NK) cells and cytotoxic T cells effectively eliminating infected cells and clearing the virus from the body. In an attempt to improve and balance the immune response, researchers introduced the human gene for the cytokine interleukin-4 (IL-4) into the mousepox virus. IL-4 stimulates B cell division and antibody synthesis, which theoretically should have led to an enhanced immune response (Clark and Pazdernik, 2016). Surprisingly, the results were contrary to expectations, as the engineered virus displayed significantly heightened virulence. It not only caused mortality in all genetically resistant mice but also claimed the lives of 50% of vaccinated mice. Excessive IL-4 expression suppressed NK cells and cytotoxic T cells while failing to enhance the antibody response (Clark and Pazdernik, 2016). Similar results have been observed with different strains of Vaccinia virus, which is utilized for smallpox vaccination. The repercussions of inserting IL-4 or other immune regulators into smallpox itself and the potential to undermine the immune response and increase virulence remain uncertain (Clark and Pazdernik, 2016). Poxviruses possess genes called cytokine response modifier (crm) genes, designed to hinder the action of NK cells and cytotoxic T cells, further complicating the assessment of smallpox’s virulence (Clark and Pazdernik, 2016). Nevertheless, genetic engineering allows for the concealment of a potentially hazardous virus within a harmless bacterium, a phenomenon already witnessed in nature when bacteriophages incorporate their genomes into bacterial chromosomes or plasmids, subsequently re-emerging to infect other hosts. Theoretically, cloning the complete genome of a small animal or plant virus into a bacterial plasmid could serve as the basis for a biological weapon, while larger viruses could be accommodated using bacterial or yeast artificial chromosomes. In the case of RNA viruses, the viral genome must first be reverse transcribed into complementary DNA (cDNA) before cloning into a bacterial vector. Viruses harboring toxic sequences, which are not stably maintained on bacterial plasmids, could potentially be cloned as separate fragments. This approach has proven successful with yellow fever virus albeit requiring in vitro ligation of the fragments to generate a complete, functional cDNA (Clark and Pazdernik, 2016). Different cell types, including bacteria and eukaryotes, are capable of taking up DNA or RNA under specific conditions through transformation. Consequently, the naked nucleic acid genomes of numerous DNA and RNA viruses retain their infectivity even without their protein capsids or envelopes. Once a viral genome is cloned, the DNA molecule containing it becomes infectious itself. Alternatively, cDNA versions of RNA viruses can infect host cells, giving rise to new virus particles containing RNA. Poliovirus, influenza, and coronavirus are among the RNA viruses that have demonstrated this capability. An ingenious strategy for generating an RNA virus involves cloning the cDNA version of its genome downstream of a strong promoter in a bacterial plasmid. Transcription within the induced bacterial cell results in the production of a large number of infectious viral particles. Combining a dangerous human RNA virus with a harmless intestinal bacterium, controlled by a promoter responsive to intestinal conditions, could pose a significant threat (Clark and Pazdernik, 2016). While certain pathogenic bacteria exhibit slow growth or are challenging to the culture outside their host organisms, advancements in biotechnology have facilitated the identification of infectious microbes through molecular diagnostics. Instead of relying on classical microbiological techniques to grow and identify disease-causing agents, molecular diagnostics employ the analysis of molecules, primarily DNA but also RNA, proteins, and volatile organic compounds. These molecular techniques offer advantages in terms of speed, accuracy, and sensitivity.
Fluorescent in situ hybridization (FISH) is one such diagnostic method, involving the direct probing of biopsies or patient samples with fluorescent DNA oligonucleotides specific to a target pathogen (Clark and Pazdernik, 2016). If the pathogen is present, the probe binds to complementary DNA in its chromosome, enabling visualization of fluorescence under a microscope. An innovative approach utilizing peptide nucleic acid (PNA), which replaces the negatively charged sugar-phosphate backbone of DNA with a neutral peptide backbone, enhances the binding affinity of PNA probes to complementary DNA and facilitates their entry into bacterial cells. PCR amplification of target DNA sequences, unique to a particular pathogen, is another commonly used method in molecular diagnostics (Clark and Pazdernik, 2016). The ability to design primers specific to a pathogen allows for the amplification of target DNA, enabling PCR to serve as a diagnostic tool. PCR’s versatility stems from its potential to detect a single molecule of target DNA and its applicability to microbes that are challenging to culture in the laboratory. However, PCR is susceptible to contamination and false positives. Randomly amplified polymorphic DNA (RAPD), a PCR variant, distinguishes different strains of the same bacterial species, aiding in epidemiological studies to track the spread of infectious diseases. Each microorganism species possesses a unique small-subunit ribosomal RNA (SSU rRNA) sequence, such as 16S rRNA in bacteria and 18S rRNA in eukaryotes (Clark and Pazdernik, 2016). Therefore, clinicians can employ PCR to amplify the gene encoding the microbe’s SSU rRNA when faced with an unknown infection. By sequencing and comparing the PCR fragment to a database of known DNA sequences, the pathogen can be identified. Checkerboard hybridization is a technique that utilizes SSU rRNA as a basis, allowing for the simultaneous detection and identification of multiple bacteria in a single sample. Horizontal lines on a hybridization membrane contain probes specific to different bacteria, while PCR amplification of the SSU rRNA gene from clinical samples, potentially harboring a mixture of pathogens, generates fluorescently labeled fragments that are applied vertically to the membrane. Fluorescent spots indicate samples that have hybridized with the probes. Abbott Laboratories has developed a potentially revolutionary technology called PLEX-ID, which combines traditional PCR with mass spectrometry to identify unknown microbes in patient samples (Clark and Pazdernik, 2016). By analyzing the mass of amplified DNA fragments using a mass spectrometer, the DNA sequence can be deduced, enabling the identification of the pathogen. PLEX-ID has the capability to provide a diagnosis within eight hours. In the future, disease diagnosis may be achievable using an “electronic nose,” a device that detects volatile organic compounds released by pathogens or by the body in certain diseased conditions (Clark and Pazdernik, 2016).
The Role of Molecular Cloning in SARS-COV 2 Treatment
B cell receptor (BCR) repertoires display remarkable sequence diversity resulting from somatic recombination and hypermutation processes occurring during B cell development. BCRs are transmembrane receptors situated on the surface of B cells, and their variable regions interact with specific antigen epitopes to initiate an immune response through antibody production (Zhou et al., 2021). This variable region shares identical gene sequences with the corresponding antibody produced by the B cell. The variability in the variable region is generated by somatic recombination of three gene segments in the heavy (H) chain locus (V, D, J) and two gene segments in the light (L) chain locus (V, J) (Zhou et al., 2021). The variable region of an antibody, also known as immunoglobulin (Ig), determines its specificity for binding to a specific viral antigenic epitope. Somatic hypermutation occurs during B cell proliferation in the germinal center, introducing random mutations in the genes encoding the variable region of individual monoclonal antibodies (mAbs) (Zhou et al., 2021). This process is crucial for the development of high-affinity antibodies, a phenomenon referred to as antibody affinity maturation. Importantly, no two B cells possess identical BCRs. Therefore, the cloning of a functional mAb requires the analysis and cloning of one B cell at a time to ensure the native pairing of antibody heavy and light chains (Bassing et al., 2002).
Advancements in antibody gene cloning techniques, such as hybridoma technology, human B cell immortalization, antibody phage display, human immunoglobulin transgenic mice, and single B cell antibody technology, have revolutionized the process of cloning functional human neutralizing antibodies (HuNAbs) (Tiller, 2011). Among these techniques, single B cell-based antibody cloning has been extensively employed to isolate SARS-CoV-2-specific antibodies during the ongoing COVID-19 pandemic. This method involves amplifying auto-paired Ig heavy- and light-chain RNA sequences from a heterogeneous population of memory B cells, followed by their in vitro construction into functional mAbs (Niu et al., 2019). The successful application of this technique has been demonstrated in the isolation of neutralizing mAbs against various viral infections, such as human immunodeficiency virus (HIV) and SARS-CoV-2 (Corti et al., 2015). In recent years, the combination of single-cell reverse transcription polymerase chain reaction (RT-PCR) and single B cell sorting has significantly improved the success rate of antibody gene cloning. The acquisition of single antigen-binding memory B cells through fluorescence-activated cell sorting (FACS) or optofluidic platforms has been a major technical advancement (Zhou et al., 2021). These techniques enable subsequent nested RT-PCR using primers designed to amplify naturally paired antibody heavy- and light-chain gene sequences from individual memory B cells (Liao et al., 2009). Furthermore, high-throughput single-cell RNA and VDJ deep sequencing of BCR repertoires, coupled with bioinformatics analysis, have surpassed single-cell RT-PCR in terms of efficiently screening large pools of diverse memory B cells (Cao et al., 2020). Notably, immunization using RBD/S proteins or direct infection of mice with genetically humanized immune systems has been demonstrated to generate complete HuNAbs against SARS-CoV-2 (Hansen et al., 2020). This platform has successfully facilitated the cloning and screening of certain SARS-CoV-2-specific HuNAbs. Deep sequencing of variable regions and BCR repertoires has provided insights into the characteristics of human antibody heavy and light chain pairing (Zhou et al., 2021). In addition, the combination of microfluidic-based techniques and bioinformatics analysis has the potential to further enhance the efficiency of identifying highly potent HuNAbs against specific viral antigens (Hansen et al., 2020). These advancements hold implications for combating COVID-19 and other emerging infectious diseases.
Phage display has emerged as a widely utilized approach for cloning human antibodies. This technique involves two main stages: constructing an antibody gene library and screening for antibodies specific to a particular antigen (Zhou et al., 2021). To generate a human Fab library, researchers can extract peripheral blood mononuclear cells (PBMCs) from individuals who have recovered from COVID-19. By amplifying the antibody gene pool using specific primers targeting the variable regions of the heavy and light chains, a phage library can be constructed (Zhou et al., 2021). The library can also be synthesized using selected human germline immunoglobulin variable segments, with diversity introduced in the complementarity-determining regions (CDRs) through mutagenesis. The library can be screened using SARS-CoV-2 receptor-binding domain (RBD) as a bait to isolate RBD-specific human Fabs (Zhou et al., 2021). These Fabs can then be further developed into full-length IgG1 antibodies for subsequent testing of their biochemical and functional properties. Although the phage display technique has some limitations, such as the unnatural pairing of heavy and light chains and a time-consuming panning procedure, it remains a valuable tool for obtaining antigen-specific antibodies, including potent SARS-CoV-2-specific human neutralizing antibodies (HuNAbs) (Zhou et al., 2021). The SARS-CoV-2 spike protein (S protein) plays a crucial role in viral entry by binding to the ACE2 receptor on host cells (Lv et al., 2020). The receptor-binding domain (RBD) within the S1 subunit of the S protein undergoes conformational changes, transitioning between “up” and “down” states. The “up” conformation is particularly targeted by HuNAbs since it represents the active state for ACE2 binding (Zhou et al., 2021). Multiple neutralizing domains within the S1 region have been identified, including RBD and the N-terminal domain (NTD). HuNAbs can be classified into three main groups based on their binding sites (Yuan et al., 2021). The first group, known as receptor-binding site (RBS) antibodies, targets epitopes within the RBD that overlap with the ACE2 binding site (Brouwer et al., 2020). RBS antibodies can further be divided into subclasses based on their interaction angles with the viral RBD (Yuan et al., 2021). The second group includes cross-reactive antibodies, such as CR3022, which recognize cryptic sites in the RBD and exhibit varying neutralizing activities against both SARS-CoV and SARS-CoV-2 (Yuan et al., 2021). The third group is represented by antibodies like S309, which engage the RBD through an epitope containing the N343 glycan and can neutralize both SARS-CoV-2 and SARS-CoV (Yuan et al., 2021).
Several anti-SARS-CoV-2 HuNAbs have been developed and undergone clinical trials. Paired HuNAbs have been formulated to enhance the breadth of neutralization against emerging SARS-CoV-2 variants and minimize the occurrence of HuNAb escape variants. HuNAb combinations under Emergency Use Authorization (EUA) exhibit live virus neutralization IC50 values below 0.1 µg/ml (Jones et al., 2021). Typically, these paired antibodies consist of one RBS-A HuNAb along with either one RBS-B or one RBS-C HuNAb. For example, the combination of LY-CoV555 (bamlanivimab, BAM) and LY-CoV016 (etesevimab, ETE) from Eli Lilly has been administered to COVID-19 patients (Zhou et al., 2021). The randomized phase 2/3 trial demonstrated a statistically significant reduction in viral load at day 11 among non-hospitalized patients with mild to moderate illness who received the combination therapy (Gottlieb et al., 2021). Similarly, the REGN-CoV2 antibody cocktail (casirivimab, CAS, and imdevimab, IMD) showed a substantial reduction in viral loads in patients with baseline viral load higher than 107 copies/mL compared to the placebo group (Hansen et al., 2020). Due to concerns regarding reduced neutralizing abilities against SARS-CoV-2 variants of concern (VOCs), ongoing clinical trials are investigating the efficacy of more broadly reactive HuNAbs, as well as next-generation antibodies such as bispecific antibodies and engineered antibodies. In addition, the development of BRII-196 and BRII-198, an antibody combination approved for clinical use in China, has shown promising results with 78% efficacy (Zhou et al., 2021). SARS-CoV-2 virus has the ability to undergo mutations during its replication in humans. Unlike the SARS virus in 2003, which was eliminated, COVID-19 has persisted, leading to concerns about viral mutations that could evade immunity from natural infection or vaccination (Romano et al., 2020). Several VOCs have emerged during the pandemic, including the Alpha, Beta, Gamma, Delta, and Omicron variants (Chakraborty et al., 2022). These VOCs have mutations in the spike (S) glycoprotein of the virus, which can confer resistance to human neutralizing antibodies (HuNAbs) and increase viral transmissibility. The D614G mutation, found in all VOCs, enhances viral transmissibility (Plante et al., 2021). Mutations such as E484K/A and amino acid deletions in the S protein reduce the potency of HuNAbs targeting specific regions (Jangra et al., 2021). The emergence of these VOCs poses challenges to public health and vaccine responses.
Efforts have been focused on developing vaccines to combat COVID-19, and several vaccines have been approved for emergency use. These include mRNA-based vaccines, adenovirus-vectored vaccines, and inactivated vaccines (Zhou et al., 2021). The efficacy rates reported for these vaccines mainly reflect their ability to prevent severe diseases, while their effectiveness against SARS-CoV-2 nasal infection and VOCs requires further investigation (Zhou et al., 2021). Studies have shown reduced neutralization of VOCs by antibodies from vaccinated individuals, indicating the potential for decreased effectiveness against certain variants (Wang et al., 2021). However, widespread vaccination campaigns have contributed to reduced hospitalizations, deaths, and infections in various countries. Future strategies for vaccination and immunotherapy should aim to provide broadly reactive immune protection against multiple variants (Zhou et al., 2021). The role of mucosal immunity and tissue-resident memory T cells in the upper respiratory tract needs to be explored for long-term protection (Baum et al., 2020). In addition, careful monitoring of antibody-mediated enhancement of SARS-CoV-2 infection and immunopathogenesis is crucial in the context of human infections and clinical management.
Synergy of Molecular Cloning and the CRISPR-Cas System in SARS-CoV-2 Treatment
COVID-19, caused by the SARS-CoV-2 virus, has triggered a global pandemic, with the emergence of highly transmissible and more fatal mutant strains. Effective management of this infectious disease requires a number of measures, including social distancing and isolation of confirmed cases. However, there is a pressing need for a highly sensitive diagnostic kit that enables rapid early detection (Lou et al., 2022). Diagnostic approaches for COVID-19 involve immunological and molecular techniques. Immunological methods detect viral antigens or antibodies in the lungs or blood, aiding in disease understanding and transmission dynamics (Mahase, 2020). Molecular methods primarily focus on nucleic acid detection, with RT-PCR being the gold standard due to its high sensitivity and accuracy (Broughton et al., 2020). However, the limited availability of RT-PCR equipment and materials can lead to delays and false negative results, making it unsuitable for mass screening (Broughton et al., 2020, Li and Ren, 2020). Hence, there is a need for simplified and time-efficient diagnostic methods with high accuracy (Xiang et al., 2020). The CRISPR-Cas system, with nucleic acid detection technologies like SHERLOCK (Cas13a), DETECTR (Cas12a), CDetection (Cas12b), and CAS14-DETECTR, offers a new avenue for pathogen screening (Ding et al., 2021). In recent times, diverse detection methods combined with isothermal amplification and the CRISPR-Cas system have emerged as rapid diagnostic tools for detecting SARS-CoV-2 viral RNA. For instance, a visual analysis method called CLAP combining AUNP and Cas12a-assisted RT-LAMP demonstrated the ability to detect low levels of SARS-CoV-2 RNA rapidly (Lou et al., 2022). In addition, engineered Cas12a enzyme-based LAMP showed promising results in detecting wild-type and mutant SARS-CoV-2 within a short time frame (Lou et al., 2022). Cas13a crRNAs can specifically target SARS-CoV-2 and related coronaviruses, and when combined with a generic autonomous enzyme-free hybridization chain reaction (HCR), they offer a detection method for monitoring virus transmission via objects (Yang et al., 2021). The CRISPR-Cas13 amplification strategy holds potential for efficient surveillance of SARS-CoV-2 transmission (Lou et al., 2022). Viral control over host cells has long intrigued virologists due to the limited number of viral genes. The explanation lies, in part, in the utilization of host factors by viruses during their life cycle. Identifying these host factors that either facilitate or impede novel virus replication can uncover potential targets for antiviral therapies (Deol et al., 2022). A number of techniques, including forward genetic screens, are employed to study virus-host interactions. Although the CRISPR/Cas9 system is not the first tool for genetic screens, it has emerged as a highly robust method. Genome-wide CRISPR screens are being conducted to identify host genes involved in SARS-CoV-2 replication. For instance, Wang et al. identified ACE-2 as the entry receptor and highlighted TMEM106B, VAC14, cholesterol regulators, and exocyst subunits as additional host factors supporting SARS-CoV-2 infection, thus offering potential targets for antiviral strategies (Wang et al., 2021). Other researchers have also employed CRISPR-based screens to identify candidate host genes for SARS-CoV-2, paving the way for the development of CRISPR/Cas9-mediated antiviral therapeutics (Daniloski et al., 2021, Zhu et al., 2021). Huang et al. devised a CRISPR-based diagnostic approach that utilizes customized CRISPR Cas12a/gRNA complex and fluorescent probes to detect target amplifiers generated by standard RT-PCR or isothermal recombinase polymerase amplification (RPA). This method enables sensitive detection at locations where the RT-PCR system necessary for qPCR diagnosis is unavailable (Huang et al., 2021). The technique exhibited high sensitivity, with a reaction time of approximately 50 minutes and a detection limit of two samples per sample.
The diagnostic results obtained using the CRISPR analysis from nasal swab samples of suspected COVID-19 patients were comparable to quantitative RT-PCR tests and outperformed standard clinical laboratory tests. With no specific treatment available for emerging infectious diseases like COVID-19, it is crucial to explore effective diagnostic and therapeutic approaches (Lou et al., 2022). A number of antiviral approaches focus on inhibiting different stages of the viral life cycle by targeting structural or non-structural components. CRISPR/Cas systems have gained significant attention as a means to limit viral replication by specifically targeting the viral genome. CRISPR/Cas9, in particular, has demonstrated its potential as an antiviral strategy against DNA viruses, both in vitro and in vivo (Lee, 2019). Similarly, the CRISPR/Cas12a system has shown promise in inactivating integrated HIV DNA genomes, surpassing Cas9 in HIV inhibition. On the contrary, the CRISPR/Cas13 system has emerged as a novel RNA-guided RNA targeting system, capable of recognizing and degrading the genomic RNA of SARS-CoV-2, thereby impeding virus replication. Recent research has demonstrated the efficacy of CRISPR-Cas13 in safeguarding host bacterial cells against phage infection through specific cRNA (Yan et al., 2019). This strategy can potentially be leveraged to design therapeutic drugs targeting the single-stranded RNA genomes of emerging infectious diseases. The Cas13 enzymes utilize a short hairpin crRNA to recognize specific sequences on the target RNA, and unlike Cas9, they do not require a protospacer adjacent motif (PAM) sequence (Burmistrz et al., 2020). Some subtypes of Cas13 also exhibit collateral cleavage activity, resulting in non-specific cleavage of target and non-target RNAs. This property has been utilized in diagnostic applications such as SHERLOCK. Different subtypes of Cas13 have been explored for their antiviral potential. Abbott et al. introduced a CRISPR-Cas13-based strategy known as PAC-MAN (Prophylactic Antiviral CRISPR in Human Cells), which effectively degrades SARS-CoV-2 RNA in cells by identifying functional crRNA specifically targeting SARS-CoV-2 (Lou et al., 2022). This method reduces viral load within cells, and a small set of six crRNAs can target over 90% of coronaviruses, making PAC-MAN a promising pan-coronavirus suppression strategy (Abbott et al., 2020). The development of CRISPR-based tools for the treatment of emerging infectious diseases holds great research prospects.
The primary drawback hindering the extensive use of the CRISPR-Cas system lies in its inability to accurately identify specific nucleic acids for diagnostic and therapeutic purposes (Doudna, 2020). To address this issue, one approach is to enhance the specificity of target nucleic acids by modifying the CRISPR protein, thus minimizing off-target effects (Naeem et al., 2020). Bioinformatics methods are commonly employed to detect off-target effects, and advancements in Cas9 have shown promise in reducing such effects (Lou et al., 2022, Coelho et al., 2020). Future research should focus on investigating off-target effects and refining detection techniques. In utilizing the CRISPR-Cas system for the prevention and control of infectious diseases, delivery tools play a crucial role in targeting cells within the body (Lou et al., 2022). However, the carrier function of CRISPR is limited by the size of viral genes, and most Cas proteins are relatively large in molecular weight (Liu et al., 2020). Overcoming this challenge necessitates the search for low molecular weight Cas proteins (Luther et al., 2018). Moreover, when Cas proteins derived from prokaryotes are administered to the human body, they can elicit an immune response and trigger the production of specific antibodies, which interfere with CRISPR’s immune response (Lou et al., 2022). Enhancing the efficiency of the CRISPR-Cas system is vital to mitigate the immune response in future CRISPR tool development. Cas9 and Cas12 proteins exhibit PAM-dependent recognition and cleavage of dsDNA, allowing them to target specific gene sequences within target genes (Karvelis et al., 2020). However, some highly specific sequences may not be accessible, emphasizing the need for further enhancement of the CRISPR tool for target selection (Lou et al., 2022). In addition, the presence of RNA enzymes widely in existence leads to RNA instability, thereby affecting the diagnostic efficacy of CRISPR.
Conclusion
There has been considerable progress in molecular cloning techniques in research laboratories as well as widespread operation in medical microbiology. While DNA-based procedures have improved the diagnosis of diseases, conventional cell factories have drained their competence (Khan et al., 2016). It is obligatory to discover and integrate new production systems into the production process. In addition, a deeper understanding of the physiology of host cells and their responses to stress would enable enhanced tools by gene manipulation either at the genetic or at the metabolic levels for growing yield and eminence (Khan et al., 2016). The progressions in recombinant DNA technology, microbiology, genomics, bioinformatics, and related fields have, however, contributed significantly to the understanding of the pathogenic mechanisms underlying the microbial infectious diseases and how pathogens interrelate with hosts. Several new vaccine strategies have been established, as outcomes of these advances have produced promising results. Masses of children, globally, still die from infectious diseases notwithstanding the existence of currently available vaccines. Therefore, it is essential to understand the trials involved in developing recombinant vaccines and estimate the stability between charge, remunerations, and hazard before bringing a vaccine applicant to the hospital. As a result of this approach, patients with unrecognized or hard-to-diagnose infections will be identified and treated punctually, resulting in reduced stays and a reduction in iatrogenic events. Considering the relative costs of novel diagnostics compared to existing standards, public reimbursements will need to be carefully explored. Hence, molecular cloning can be delineated as a scientific effort to treat a lengthy list of diseases that have claimed a lot of human lives in the course of long period.
References
Abbott, T. R., Dhamdhere, G., Liu, Y., Lin, X., Goudy, L., Zeng, L., Chemparathy, A., Chmura, S., Heaton, N. S., Debs, R., Pande, T., Endy, D., La Russa, M. F., Lewis, D. B., & Qi, L. S. (2020). Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell, 181(4), 865–876.e12. https://doi.org/10.1016/j.cell.2020.04.020https://doi.org/10.1016/j.cell.2020.04.020
Adrio, J. L., & Demain, A. L. (2010). Recombinant organisms for production of industrial products. Bioengineered Bugs, 1(2), 116–131. https://doi.org/10.4161/bbug.1.2.10484https://doi.org/10.4161/bbug.1.2.10484
Ahmad, M., Hirz, M., Pichler, H., & Schwab, H. (2014). Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Applied Microbiology and Biotechnology, 98(12), 5301–5317. https://doi.org/10.1007/s00253-014-5732-5https://doi.org/10.1007/s00253-014-5732-5
Aiuti, A., & Roncarolo, M. G. (2009). Ten years of gene therapy for primary immune deficiencies. Hematology. American Society of Hematology. Education Program, 682–689. https://doi.org/10.1182/asheducation-2009.1.682https://doi.org/10.1182/asheducation-2009.1.682
Aiuti, A., Vai, S., Mortellaro, A., Casorati, G., Ficara, F., Andolfi, G., Ferrari, G., Tabucchi, A., Carlucci, F., Ochs, H. D., Notarangelo, L. D., Roncarolo, M. G., & Bordignon, C. (2002). Immune reconstitution in ADA-SCID after PBL gene therapy and discontinuation of enzyme replacement. Nature Medicine, 8(5), 423–425. https://doi.org/10.1038/nm0502-423https://doi.org/10.1038/nm0502-423
Ali, H., & Murrell, J. C. (2009). Development and validation of promoter-probe vectors for the study of methane monooxygenase gene expression in Methylococcus capsulatus Bath. Microbiology (Reading, England), 155(Pt 3), 761–771. https://doi.org/10.1099/mic.0.021816-0https://doi.org/10.1099/mic.0.021816-0
Alibek, K. (1999). with Stephen Handelman. Biohazard. chrome-extension://efaidnbmnnnibpcajpcglclefindmkaj/https://www.nlm.nih.gov/nichsr/esmallpox/biohazard_alibek.pdfhttps://www.nlm.nih.gov/nichsr/esmallpox/biohazard_alibek.pdf
Amann, R. I., Ludwig, W., & Schleifer, K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiological Reviews, 59(1), 143–169. https://doi.org/10.1128/mr.59.1.143-169.1995https://doi.org/10.1128/mr.59.1.143-169.1995
Bandaranayake, A. D., Correnti, C., Ryu, B. Y., Brault, M., Strong, R. K., & Rawlings, D. J. (2011). Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors. Nucleic acids Research, 39(21), e143. https://doi.org/10.1093/nar/gkr706https://doi.org/10.1093/nar/gkr706
Bassing, C. H., Swat, W., & Alt, F. W. (2002). The mechanism and regulation of chromosomal V(D)J recombination. Cell, 109 Suppl, S45–S55. https://doi.org/10.1016/s0092-8674(02)00675-xhttps://doi.org/10.1016/s0092-8674(02)00675-x
Baum, A., Fulton, B. O., Wloga, E., Copin, R., Pascal, K. E., Russo, V., Giordano, S., Lanza, K., Negron, N., Ni, M., Wei, Y., Atwal, G. S., Murphy, A. J., Stahl, N., Yancopoulos, G. D., & Kyratsous, C. A. (2020). Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science (New York, N.Y.), 369(6506), 1014–1018. https://doi.org/10.1126/science.abd0831https://doi.org/10.1126/science.abd0831
Bermúdez-Humarán, L. G., Kharrat, P., Chatel, J. M., & Langella, P. (2011). Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microbial Cell Factories, 10(Suppl 1), S4. https://doi.org/10.1186/1475-2859-10-S1-S4https://doi.org/10.1186/1475-2859-10-S1-S4
Broughton, J. P., Deng, X., Yu, G., Fasching, C. L., Servellita, V., Singh, J., Miao, X., Streithorst, J. A., Granados, A., Sotomayor-Gonzalez, A., Zorn, K., Gopez, A., Hsu, E., Gu, W., Miller, S., Pan, C. Y., Guevara, H., Wadford, D. A., Chen, J. S., & Chiu, C. Y. (2020). CRISPR-Cas12-based detection of SARS-CoV-2. Nature Biotechnology, 38(7), 870–874. https://doi.org/10.1038/s41587-020-0513-4https://doi.org/10.1038/s41587-020-0513-4
Brouwer, P. J. M., Caniels, T. G., van der Straten, K., Snitselaar, J. L., Aldon, Y., Bangaru, S., Torres, J. L., Okba, N. M. A., Claireaux, M., Kerster, G., Bentlage, A. E. H., van Haaren, M. M., Guerra, D., Burger, J. A., Schermer, E. E., Verheul, K. D., van der Velde, N., van der Kooi, A., van Schooten, J., van Breemen, M. J., … van Gils, M. J. (2020). Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science (New York, N.Y.), 369(6504), 643–650. https://doi.org/10.1126/science.abc5902https://doi.org/10.1126/science.abc5902
Buchholz, F., Angrand, P. O., & Stewart, A. F. (1998). Improved properties of FLP recombinase evolved by cycling mutagenesis. Nature Biotechnology, 16(7), 657–662. https://doi.org/10.1038/nbt0798-657https://doi.org/10.1038/nbt0798-657
Burmistrz, M., Krakowski, K., & Krawczyk-Balska, A. (2020). RNA-Targeting CRISPR-Cas Systems and Their Applications. International Journal of Molecular Sciences, 21(3), 1122. https://doi.org/10.3390/ijms21031122https://doi.org/10.3390/ijms21031122
Cao, Y., Su, B., Guo, X., Sun, W., Deng, Y., Bao, L., Zhu, Q., Zhang, X., Zheng, Y., Geng, C., Chai, X., He, R., Li, X., Lv, Q., Zhu, H., Deng, W., Xu, Y., Wang, Y., Qiao, L., Tan, Y., … Xie, X. S. (2020). Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell, 182(1), 73–84.e16. https://doi.org/10.1016/j.cell.2020.05.025https://doi.org/10.1016/j.cell.2020.05.025
Cartier, N., Hacein-Bey-Abina, S., Bartholomae, C. C., Veres, G., Schmidt, M., Kutschera, I., Vidaud, M., Abel, U., Dal-Cortivo, L., Caccavelli, L., Mahlaoui, N., Kiermer, V., Mittelstaedt, D., Bellesme, C., Lahlou, N., Lefrère, F., Blanche, S., Audit, M., Payen, E., Leboulch, P., … Aubourg, P. (2009). Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science (New York, N.Y.), 326(5954), 818–823. https://doi.org/10.1126/science.1171242https://doi.org/10.1126/science.1171242
Cavazzana-Calvo, M., Hacein-Bey, S., de Saint Basile, G., Gross, F., Yvon, E., Nusbaum, P., Selz, F., Hue, C., Certain, S., Casanova, J. L., Bousso, P., Deist, F. L., & Fischer, A. (2000). Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science (New York, N.Y.), 288(5466), 669–672. https://doi.org/10.1126/science.288.5466.669https://doi.org/10.1126/science.288.5466.669
Chakraborty, C., Bhattacharya, M., & Sharma, A. R. (2022). Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: Their significant mutations in S‐glycoprotein, infectivity, re‐infectivity, immune escape and vaccines activity. Reviews in Medical Virology, 32(2), e2270. https://doi.org/10.1002/rmv.2270https://doi.org/10.1002/rmv.2270
Chehab F. F. (1993). Molecular diagnostics: past, present, and future. Human Mutation, 2(5), 331–337. https://doi.org/10.1002/humu.1380020502https://doi.org/10.1002/humu.1380020502
Clark, D. P., & Pazdernik, N. J. (2016). Biological warfare: infectious disease and bioterrorism. Biotechnology, 687–719. https://doi.org/10.1016/B978-0-12-385015-7.00022-3https://doi.org/10.1016/B978-0-12-385015-7.00022-3
Coelho, M. A., De Braekeleer, E., Firth, M., Bista, M., Lukasiak, S., Cuomo, M. E., & Taylor, B. J. M. (2020). CRISPR GUARD protects off-target sites from Cas9 nuclease activity using short guide RNAs. Nature Communications, 11(1), 4132. https://doi.org/10.1038/s41467-020-17952-5https://doi.org/10.1038/s41467-020-17952-5
Copeland, N. G., Jenkins, N. A., & Court, D. L. (2001). Recombineering: a powerful new tool for mouse functional genomics. Nature Reviews Genetics, 2(10), 769–779. https://doi.org/10.1038/35093556https://doi.org/10.1038/35093556
Corti, D., Zhao, J., Pedotti, M., Simonelli, L., Agnihothram, S., Fett, C., Fernandez-Rodriguez, B., Foglierini, M., Agatic, G., Vanzetta, F., Gopal, R., Langrish, C. J., Barrett, N. A., Sallusto, F., Baric, R. S., Varani, L., Zambon, M., Perlman, S., & Lanzavecchia, A. (2015). Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proceedings of the National Academy of Sciences of the United States of America, 112(33), 10473–10478. https://doi.org/10.1073/pnas.1510199112https://doi.org/10.1073/pnas.1510199112
Cox, D. B., Platt, R. J., & Zhang, F. (2015). Therapeutic genome editing: prospects and challenges. Nature Medicine, 21(2), 121–131. https://doi.org/10.1038/nm.3793https://doi.org/10.1038/nm.3793
Dangi, A. K., Sinha, R., Dwivedi, S., Gupta, S. K., & Shukla, P. (2018). Cell line techniques and gene editing tools for antibody production: A review. Frontiers in Pharmacology, 9, 630. https://doi.org/10.3389/fphar.2018.00630https://doi.org/10.3389/fphar.2018.00630
Daniloski, Z., Jordan, T. X., Wessels, H. H., Hoagland, D. A., Kasela, S., Legut, M., Maniatis, S., Mimitou, E. P., Lu, L., Geller, E., Danziger, O., Rosenberg, B. R., Phatnani, H., Smibert, P., Lappalainen, T., tenOever, B. R., & Sanjana, N. E. (2021). Identification of required host factors for SARS-CoV-2 infection in human cells. Cell, 184(1), 92–105.e16. https://doi.org/10.1016/j.cell.2020.10.030https://doi.org/10.1016/j.cell.2020.10.030
Datta, N. (2023). A review of molecular biology detection methods for human adenovirus. AIMS Biophysics, 10(1): 95–120. doi: 10.3934/biophy.202300810.3934/biophy.2023008
De Beenhouwer, H., Lhiang, Z., Jannes, G., Mijs, W., Machtelinckx, L., Rossau, R., Traore, H., & Portaels, F. (1995). Rapid detection of rifampicin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tubercle and Lung Disease: The Official Journal of the International Union against Tuberculosis and Lung Disease, 76(5), 425–430. https://doi.org/10.1016/0962-8479(95)90009-8https://doi.org/10.1016/0962-8479(95)90009-8
Demeter, L., & Haubrich, R. (2001). Phenotypic and genotypic resistance assays: methodology, reliability, and interpretations. JAIDS Journal of Acquired Immune Deficiency Syndromes, 26, S3–S9. doi: 10.1097/00042560-200103011-0000210.1097/00042560-200103011-00002
Deol, P., Madhwal, A., Sharma, G., Kaushik, R., & Malik, Y. S. (2022). CRISPR use in diagnosis and therapy for COVID-19. Methods in Microbiology, 50, 123–150. https://doi.org/10.1016/bs.mim.2022.03.002https://doi.org/10.1016/bs.mim.2022.03.002
Descotes J. (2009). Immunotoxicity of monoclonal antibodies. MAbs, 1(2), 104–111. https://doi.org/10.4161/mabs.1.2.7909https://doi.org/10.4161/mabs.1.2.7909
Ding, R., Long, J., Yuan, M., Jin, Y., Yang, H., Chen, M., Chen, S., & Duan, G. (2021). CRISPR/Cas system: A potential technology for the prevention and control of COVID-19 and emerging infectious diseases. Frontiers in Cellular and Infection Microbiology, 11, 639108. https://doi.org/10.3389/fcimb.2021.639108https://doi.org/10.3389/fcimb.2021.639108
Doron, S., & Gorbach, S. L. (2008). Bacterial infections: Overview. International Encyclopedia of Public Health, 273–282. https://doi.org/10.1016/B978-012373960-5.00596-7https://doi.org/10.1016/B978-012373960-5.00596-7
Doudna J. A. (2020). The promise and challenge of therapeutic genome editing. Nature, 578(7794), 229–236. https://doi.org/10.1038/s41586-020-1978-5https://doi.org/10.1038/s41586-020-1978-5
Doudna, J. A., & Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science (New York, N.Y.), 346(6213), 1258096. https://doi.org/10.1126/science.1258096https://doi.org/10.1126/science.1258096
Elnifro, E. M., Ashshi, A. M., Cooper, R. J., & Klapper, P. E. (2000). Multiplex PCR: optimization and application in diagnostic virology. Clinical Microbiology Reviews, 13(4), 559–570. https://doi.org/10.1128/CMR.13.4.559https://doi.org/10.1128/CMR.13.4.559
Engler, C., Kandzia, R., & Marillonnet, S. (2008). A one pot, one step, precision cloning method with high throughput capability. PloS One, 3(11), e3647. https://doi.org/10.1371/journal.pone.0003647https://doi.org/10.1371/journal.pone.0003647
Espy, M. J., Uhl, J. R., Sloan, L. M., Rosenblatt, J. E., Cockerill, F. R., 3rd, & Smith, T. F. (2002). Detection of vaccinia virus, herpes simplex virus, varicella-zoster virus, and Bacillus anthracis DNA by LightCycler polymerase chain reaction after autoclaving: implications for biosafety of bioterrorism agents. Mayo Clinic Proceedings, 77(7), 624–628. https://doi.org/10.4065/77.7.624https://doi.org/10.4065/77.7.624
Fakruddin, M., Mazumdar, R. M., Khanjada, S. B. M., Chowdhury, A., & Hossain, M. N. (2013). Critical factors affecting the success of cloning, expression, and mass production of enzymes by recombinant E. coli. ISRN Biotechnology, 2013, 1–7. https://doi.org/10.5402/2013/590587https://doi.org/10.5402/2013/590587
Gibson, D. G., Glass, J. I., Lartigue, C., Noskov, V. N., Chuang, R. Y., Algire, M. A., Benders, G. A., Montague, M. G., Ma, L., Moodie, M. M., Merryman, C., Vashee, S., Krishnakumar, R., Assad-Garcia, N., Andrews-Pfannkoch, C., Denisova, E. A., Young, L., Qi, Z. Q., Segall-Shapiro, T. H., Calvey, C. H., … Venter, J. C. (2010). Creation of a bacterial cell controlled by a chemically synthesized genome. Science (New York, N.Y.), 329(5987), 52–56. https://doi.org/10.1126/science.1190719https://doi.org/10.1126/science.1190719
Gibson, D. G., Young, L., Chuang, R. Y., Venter, J. C., Hutchison, C. A., 3rd, & Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods, 6(5), 343–345. https://doi.org/10.1038/nmeth.1318https://doi.org/10.1038/nmeth.1318
Gottlieb, R. L., Nirula, A., Chen, P., Boscia, J., Heller, B., Morris, J., Huhn, G., Cardona, J., Mocherla, B., Stosor, V., Shawa, I., Kumar, P., Adams, A. C., Van Naarden, J., Custer, K. L., Durante, M., Oakley, G., Schade, A. E., Holzer, T. R., Ebert, P. J., … Skovronsky, D. M. (2021). Effect of Bamlanivimab as monotherapy or in combination with Etesevimab on viral load in patients with mild to moderate COVID-19: A randomized clinical trial. JAMA, 325(7), 632–644. https://doi.org/10.1001/jama.2021.0202https://doi.org/10.1001/jama.2021.0202
Gupta, S. K., & Shukla, P. (2017). Sophisticated cloning, fermentation, and purification technologies for an enhanced therapeutic protein production: A review. Frontiers in Pharmacology, 8, 419. https://doi.org/10.3389/fphar.2017.00419https://doi.org/10.3389/fphar.2017.00419
Hansen, J., Baum, A., Pascal, K. E., Russo, V., Giordano, S., Wloga, E., Fulton, B. O., Yan, Y., Koon, K., Patel, K., Chung, K. M., Hermann, A., Ullman, E., Cruz, J., Rafique, A., Huang, T., Fairhurst, J., Libertiny, C., Malbec, M., Lee, W. Y., … Kyratsous, C. A. (2020). Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science (New York, N.Y.), 369(6506), 1010–1014. https://doi.org/10.1126/science.abd0827https://doi.org/10.1126/science.abd0827
Hansen, J., Baum, A., Pascal, K. E., Russo, V., Giordano, S., Wloga, E., Fulton, B. O., Yan, Y., Koon, K., Patel, K., Chung, K. M., Hermann, A., Ullman, E., Cruz, J., Rafique, A., Huang, T., Fairhurst, J., Libertiny, C., Malbec, M., Lee, W. Y., … Kyratsous, C. A. (2020). Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science (New York, N.Y.), 369(6506), 1010–1014. https://doi.org/10.1126/science.abd0827https://doi.org/10.1126/science.abd0827
Hendrix, C. E., and Rohde, E. R. (2021). Molecular Diagnostics in the Medical Laboratory in Real Time. https://asm.org/Articles/2021/July/Molecular-Diagnostics-in-the-Medical-Laboratory-in.https://asm.org/Articles/2021/July/Molecular-Diagnostics-in-the-Medical-Laboratory-in
House, D., Chinh, N. T., Diep, T. S., Parry, C. M., Wain, J., Dougan, G., White, N. J., Hien, T. T., & Farrar, J. J. (2005). Use of paired serum samples for serodiagnosis of typhoid fever. Journal of Clinical Microbiology, 43(9), 4889–4890. https://doi.org/10.1128/JCM.43.9.4889-4890.2005https://doi.org/10.1128/JCM.43.9.4889-4890.2005
Huang, H., Patel, D. D., & Manton, K. G. (2005). The immune system in aging: roles of cytokines, T cells and NK cells. Frontiers in Bioscience: A Journal and Virtual Library, 10, 192–215. https://doi.org/10.2741/1521https://doi.org/10.2741/1521
Huang, Z., Tian, D., Liu, Y., Lin, Z., Lyon, C. J., Lai, W., Fusco, D., Drouin, A., Yin, X., Hu, T., & Ning, B. (2020). Ultra-sensitive and high-throughput CRISPR-powered COVID-19 diagnosis. Biosensors & Bioelectronics, 164, 112316. https://doi.org/10.1016/j.bios.2020.112316https://doi.org/10.1016/j.bios.2020.112316
Hutchison, C. A., 3rd, Chuang, R. Y., Noskov, V. N., Assad-Garcia, N., Deerinck, T. J., Ellisman, M. H., Gill, J., Kannan, K., Karas, B. J., Ma, L., Pelletier, J. F., Qi, Z. Q., Richter, R. A., Strychalski, E. A., Sun, L., Suzuki, Y., Tsvetanova, B., Wise, K. S., Smith, H. O., Glass, J. I., … Venter, J. C. (2016). Design and synthesis of a minimal bacterial genome. Science (New York, N.Y.), 351(6280), aad6253. https://doi.org/10.1126/science.aad6253https://doi.org/10.1126/science.aad6253
Ieven, M., Ursi, D., Van Bever, H., Quint, W., Niesters, H. G., & Goossens, H. (1996). Detection of Mycoplasma pneumoniae by two polymerase chain reactions and role of M. pneumoniae in acute respiratory tract infections in pediatric patients. The Journal of Infectious Diseases, 173, 1445–1452. https://doi.org/10.1093/infdis/173.6.1445https://doi.org/10.1093/infdis/173.6.1445
Ingham, A. B., & Moore, R. J. (2007). Recombinant production of antimicrobial peptides in heterologous microbial systems. Biotechnology and Applied Biochemistry, 47(1), 1–9.
Ingham, A. B., & Moore, R. J. (2007). Recombinant production of antimicrobial peptides in heterologous microbial systems. Biotechnology and Applied Biochemistry, 47(Pt 1), 1–9. https://doi.org/10.1042/BA20060207https://doi.org/10.1042/BA20060207
Jangra, S., Ye, C., Rathnasinghe, R., Stadlbauer, D., Personalized Virology Initiative study group, Krammer, F., Simon, V., Martinez-Sobrido, L., García-Sastre, A., & Schotsaert, M. (2021). SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. The Lancet Microbe, 2(7), e283–e284. https://doi.org/10.1016/S2666-5247(21)00068-9https://doi.org/10.1016/S2666-5247(21)00068-9
Jones, B. E., Brown-Augsburger, P. L., Corbett, K. S., Westendorf, K., Davies, J., Cujec, T. P., Wiethoff, C. M., Blackbourne, J. L., Heinz, B. A., Foster, D., Higgs, R. E., Balasubramaniam, D., Wang, L., Zhang, Y., Yang, E. S., Bidshahri, R., Kraft, L., Hwang, Y., Žentelis, S., Jepson, K. R., … Falconer, E. (2021). The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Science Translational Medicine, 13(593), eabf1906. https://doi.org/10.1126/scitranslmed.abf1906https://doi.org/10.1126/scitranslmed.abf1906
Juliane C. Lessard, (2013), Chapter Seven - Molecular Cloning, Editor(s): Jon Lorsch, Methods in Enzymology, Academic Press, Volume 529, Pages 85–98. Retrieved from https://www.sciencedirect.com/https://www.sciencedirect.com/
Kan, Y. W., & Dozy, A. M. (1978). Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proceedings of the National Academy of Sciences of the United States of America, 75(11), 5631–5635. https://doi.org/10.1073/pnas.75.11.5631https://doi.org/10.1073/pnas.75.11.5631
Karvelis, T., Bigelyte, G., Young, J. K., Hou, Z., Zedaveinyte, R., Budre, K., Paulraj, S., Djukanovic, V., Gasior, S., Silanskas, A., Venclovas, Č., & Siksnys, V. (2020). PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Research, 48(9), 5016–5023. https://doi.org/10.1093/nar/gkaa208https://doi.org/10.1093/nar/gkaa208
Khan, S., Ullah, M. W., Siddique, R., Nabi, G., Manan, S., Yousaf, M., & Hou, H. (2016). Role of Recombinant DNA Technology to Improve Life. International Journal of Genomics, 2016, 2405954. https://doi.org/10.1155/2016/2405954.https://doi.org/10.1155/2016/2405954
Kieback, D. G., Delvoux, B., Romano, A., Ollig, S., & Fischer, D. C. (2008). Persistent adenovirus-mediated thymidine kinase gene expression in ovarian cancer cells increases cell killing efficacy over time. Anticancer Research, 28(5A), 2569–2575.
Kim, H., Yoo, S. J., & Kang, H. A. (2015). Yeast synthetic biology for the production of recombinant therapeutic proteins. FEMS Yeast Research, 15(1), 1–16. https://doi.org/10.1111/1567-1364.12195https://doi.org/10.1111/1567-1364.12195
Kim, J. Y., Sajjan, U. S., Krasan, G. P., & LiPuma, J. J. (2005). Disruption of tight junctions during traversal of the respiratory epithelium by Burkholderia cenocepacia. Infection and Immunity, 73, 7107–7112. https://doi.org/10.1128/IAI.73.11.7107-7112.2005https://doi.org/10.1128/IAI.73.11.7107-7112.2005
Klarenbeek, N. B., Güler-Yüksel, M., van der Kooij, S. M., Han, K. H., Ronday, H. K., Kerstens, P. J., Seys, P. E., Huizinga, T. W., Dijkmans, B. A., & Allaart, C. F. (2011). The impact of four dynamic, goal-steered treatment strategies on the 5-year outcomes of rheumatoid arthritis patients in the BeSt study. Annals of the Rheumatic Diseases, 70(6), 1039–1046. https://doi.org/10.1136/ard.2010.141234https://doi.org/10.1136/ard.2010.141234
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., & Joung, J. K. (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature, 529(7587), 490–495. https://doi.org/10.1038/nature16526https://doi.org/10.1038/nature16526
Kommedal, O., Kvello, K., Skjastad, R., Langeland, N., & Wiker, H. G. (2009). Direct 16S rRNA gene sequencing from clinical specimens, with special focus on polybacterial samples and interpretation of mixed DNA chromatograms. Journal of Medical Microbiology, 47, 3562–3568. https://doi.org/10.1128/JCM.00973-09https://doi.org/10.1128/JCM.00973-09
Kosuri, S., & Church, G. M. (2014). Large-scale de novo DNA synthesis: technologies and applications. Nature Methods, 11(5), 499–507. https://doi.org/10.1038/nmeth.2918https://doi.org/10.1038/nmeth.2918
Kuhnel, B., Alcantara, J., Boothe, J., van Rooijen, G., & Moloney, M. (2003). Precise and efficient cleavage of recombinant fusion proteins using mammalian aspartic proteases. Protein Engineering Design and Selection, 16, 777–783. https://doi.org/10.1093/protein/gzg091https://doi.org/10.1093/protein/gzg091
Lam, P., Khan, G., Stripecke, R., Hui, K. M., Kasahara, N., Peng, K. W., & Guinn, B. A. (2013). The innovative evolution of cancer gene and cellular therapies. Cancer Gene Therapy, 20(3), 141–149. https://doi.org/10.1038/cgt.2012.93https://doi.org/10.1038/cgt.2012.93
Lee C. (2019). CRISPR/Cas9-Based Antiviral Strategy: Current Status and the Potential Challenge. Molecules (Basel, Switzerland), 24(7), 1349. https://doi.org/10.3390/molecules24071349https://doi.org/10.3390/molecules24071349
Li Y. (2009). Carrier proteins for fusion expression of antimicrobial peptides in Escherichia coli. Biotechnology and Applied Biochemistry, 54(1), 1–9. https://doi.org/10.1042/BA20090087https://doi.org/10.1042/BA20090087
Li, C., & Ren, L. (2020). Recent progress on the diagnosis of 2019 Novel Coronavirus. Transboundary and Emerging Diseases, 67(4), 1485–1491. https://doi.org/10.1111/tbed.13620https://doi.org/10.1111/tbed.13620
Li, M. Z., & Elledge, S. J. (2007). Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nature Methods, 4(3), 251–256. https://doi.org/10.1038/nmeth1010https://doi.org/10.1038/nmeth1010
Liao, H. X., Levesque, M. C., Nagel, A., Dixon, A., Zhang, R., Walter, E., Parks, R., Whitesides, J., Marshall, D. J., Hwang, K. K., Yang, Y., Chen, X., Gao, F., Munshaw, S., Kepler, T. B., Denny, T., Moody, M. A., & Haynes, B. F. (2009). High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. Journal of Virological Methods, 158(1–2), 171–179. https://doi.org/10.1016/j.jviromet.2009.02.014https://doi.org/10.1016/j.jviromet.2009.02.014
Lin, C. H., Pan, Y. C., Liu, F. W., & Chen, C. Y. (2017). Prokaryotic expression and action mechanism of antimicrobial LsGRP1C recombinant protein containing a fusion partner of small ubiquitin-like modifier. Applied Microbiology and Biotechnology, 101(22), 8129–8138. https://doi.org/10.1007/s00253-017-8530-zhttps://doi.org/10.1007/s00253-017-8530-z
Liu, Q., Zhang, H., & Huang, X. (2020). Anti-CRISPR proteins targeting the CRISPR-Cas system enrich the toolkit for genetic engineering. The FEBS Journal, 287(4), 626–644. https://doi.org/10.1111/febs.15139https://doi.org/10.1111/febs.15139
Lou, J., Wang, B., Li, J. et al. (2022). The CRISPR-Cas system as a tool for diagnosing and treating infectious diseases. Mol Biol Rep 49, 11301–11311. https://doi.org/10.1007/s11033-022-07752-zhttps://doi.org/10.1007/s11033-022-07752-z
Lu, J. Y., Lin, Y. Y., Qian, J., Tao, S. C., Zhu, J., Pickart, C., & Zhu, H. (2008). Functional dissection of a HECT ubiquitin E3 ligase. Molecular and Cellular Proteomics, 7, 35–45. https://doi.org/10.1074/mcp.M700353-MCP200https://doi.org/10.1074/mcp.M700353-MCP200
Luan, C., Zhang, H. W., Song, D. G., Xie, Y. G., Feng, J., & Wang, Y. Z. (2014). Expressing antimicrobial peptide cathelicidin-BF in Bacillus subtilis using SUMO technology. Applied Microbiology and Biotechnology, 98(8), 3651–3658. https://doi.org/10.1007/s00253-013-5246-6https://doi.org/10.1007/s00253-013-5246-6
Luther, D. C., Lee, Y. W., Nagaraj, H., Scaletti, F., & Rotello, V. M. (2018). Delivery approaches for CRISPR/Cas9 therapeutics in vivo: advances and challenges. Expert Opinion on Drug Delivery, 15(9), 905–913. https://doi.org/10.1080/17425247.2018.1517746https://doi.org/10.1080/17425247.2018.1517746
Lv, Z., Deng, Y. Q., Ye, Q., Cao, L., Sun, C. Y., Fan, C., Huang, W., Sun, S., Sun, Y., Zhu, L., Chen, Q., Wang, N., Nie, J., Cui, Z., Zhu, D., Shaw, N., Li, X. F., Li, Q., Xie, L., Wang, Y., … Wang, X. (2020). Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science (New York, N.Y.), 369(6510), 1505–1509. https://doi.org/10.1126/science.abc5881https://doi.org/10.1126/science.abc5881
Mahase E. (2020). Covid-19: Coronavirus was first described in The BMJ in 1965. BMJ (Clinical Research Ed.), 369, m1547. https://doi.org/10.1136/bmj.m1547https://doi.org/10.1136/bmj.m1547
Marttila, H. J., Soini, H., Vyshnevskaya, E., Vyshnevskiy, B. I., Otten, T. F., Vasilyef, A. V., & Viljanen, M. K. (1999). Line probe assay in the rapid detection of rifampin-resistant Mycobacterium tuberculosis directly from clinical specimens. Scandinavian Journal of Infectious Diseases, 31(3), 269–273. https://doi.org/10.1080/00365549950163563https://doi.org/10.1080/00365549950163563
Meiyalaghan, S., Latimer, J. M., Kralicek, A. V., Shaw, M. L., Lewis, J. G., Conner, A. J., & Barrell, P. J. (2014). Expression and purification of the antimicrobial peptide GSL1 in bacteria for raising antibodies. BMC Research Notes, 7, 777. https://doi.org/10.1186/1756-0500-7-777https://doi.org/10.1186/1756-0500-7-777
Meng, D. M., Zhao, J. F., Ling, X., Dai, H. X., Guo, Y. J., Gao, X. F., Dong, B., Zhang, Z. Q., Meng, X., & Fan, Z. C. (2017). Recombinant expression, purification and antimicrobial activity of a novel antimicrobial peptide PaDef in Pichia pastoris. Protein Expression and Purification, 130, 90–99. https://doi.org/10.1016/j.pep.2016.10.003https://doi.org/10.1016/j.pep.2016.10.003
Milly, T. A., & Tal‐Gan, Y. (2023). Targeting peptide‐based quorum sensing systems for the treatment of gram‐positive bacterial infections. Peptide Science, 115(2), e24298.
Miyake, K., Iijima, O., Suzuki, N., Matsukura, M., & Shimada, T. (2001). Selective killing of human immunodeficiency virus-infected cells by targeted gene transfer and inducible gene expression using a recombinant human immunodeficiency virus vector. Human Gene Therapy, 12, 227–233. https://doi.org/10.1089/10430340150218378https://doi.org/10.1089/10430340150218378
Montini, E., Biffi, A., Calabria, A., Biasco, L., Cesani, M., Benedicenti, F., … & Naldini, L. (2012). Integration site analysis in a clinical trial of lentiviral vector based hematopoietic stem cell gene therapy for metachromatic leukodystrophy. In Human Gene Therapy (Vol. 23, No. 10, pp. A13-A14). 140
Morgan, R. A., Dudley, M. E., Wunderlich, J. R., Hughes, M. S., Yang, J. C., Sherry, R. M., Royal, R. E., Topalian, S. L., Kammula, U. S., Restifo, N. P., Zheng, Z., Nahvi, A., de Vries, C. R., Rogers-Freezer, L. J., Mavroukakis, S. A., & Rosenberg, S. A. (2006). Cancer regression in patients after transfer of genetically engineered lymphocytes. Science (New York, N.Y.), 314(5796), 126–129. https://doi.org/10.1126/science.1129003https://doi.org/10.1126/science.1129003
Müller, C., Nolden, S., Gebhardt, P., Heinzelmann, E., Lange, C., Puk, O., Welzel, K., Wohlleben, W., & Schwartz, D. (2007). Sequencing and analysis of the biosynthetic gene cluster of the lipopeptide antibiotic Friulimicin in Actinoplanes friuliensis. Antimicrobial Agents and Chemotherapy, 51(3), 1028–1037. https://doi.org/10.1128/AAC.00942-06https://doi.org/10.1128/AAC.00942-06
Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G., & Erlich, H. (1986). Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symposia on Quantitative Biology, 51 Pt 1, 263–273. https://doi.org/10.1101/sqb.1986.051.01.032https://doi.org/10.1101/sqb.1986.051.01.032
Muriana, P. M., & Klaenhammer, T. R. (1991). Cloning, phenotypic expression, and DNA sequence of the gene for lactacin F, an antimicrobial peptide produced by Lactobacillus spp. Journal of Bacteriology, 173(5), 1779–1788.
Naeem, M., Majeed, S., Hoque, M. Z., & Ahmad, I. (2020). Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing. Cells, 9(7), 1608. https://doi.org/10.3390/cells9071608https://doi.org/10.3390/cells9071608
Nguyen, H., Martinez, B., Oganesyan, N., & Kim, R. (2004). An automated small-scale protein expression and purification screening provides beneficial information for protein production. Journal of Structural and Functional Genomics, 5, 23–27. https://doi.org/10.1023/B:JSFG.0000029195.73810.86https://doi.org/10.1023/B:JSFG.0000029195.73810.86
Nishimasu, H., Ran, F. A., Hsu, P. D., Konermann, S., Shehata, S. I., Dohmae, N., Ishitani, R., Zhang, F., & Nureki, O. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell, 156(5), 935–949. https://doi.org/10.1016/j.cell.2014.02.001https://doi.org/10.1016/j.cell.2014.02.001
Niu, X., Zhao, L., Qu, L., Yao, Z., Zhang, F., Yan, Q., Zhang, S., Liang, R., Chen, P., Luo, J., Xu, W., Lv, H., Liu, X., Lei, H., Yi, C., Li, P., Wang, Q., Wang, Y., Yu, L., Zhang, X., … Chen, L. (2019). Convalescent patient-derived monoclonal antibodies targeting different epitopes of E protein confer protection against Zika virus in a neonatal mouse model. Emerging Microbes & Infections, 8(1), 749–759. https://doi.org/10.1080/22221751.2019.1614885https://doi.org/10.1080/22221751.2019.1614885
Nizet V. (2006). Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Current Issues in Molecular Biology, 8(1), 11–26.
Nurk, S., Koren, S., Rhie, A., Rautiainen, M., Bzikadze, A. V., Mikheenko, A., Vollger, M. R., Altemose, N., Uralsky, L., Gershman, A., Aganezov, S., Hoyt, S. J., Diekhans, M., Logsdon, G. A., Alonge, M., Antonarakis, S. E., Borchers, M., Bouffard, G. G., Brooks, S. Y., Caldas, G. V., … Phillippy, A. M. (2022). The complete sequence of a human genome. Science (New York, N.Y.), 376(6588), 44–53. https://doi.org/10.1126/science.abj6987https://doi.org/10.1126/science.abj6987
O’Dell J. R. (2004). Therapeutic strategies for rheumatoid arthritis. The New England Journal of Medicine, 350(25), 2591–2602. https://doi.org/10.1056/NEJMra040226https://doi.org/10.1056/NEJMra040226
Orrapin, S., & Intorasoot, S. (2014). Recombinant expression of novel protegrin-1 dimer and LL-37-linker-histatin-5 hybrid peptide mediated biotin carboxyl carrier protein fusion partner. Protein Expression and Purification, 93, 46–53. https://doi.org/10.1016/j.pep.2013.10.010https://doi.org/10.1016/j.pep.2013.10.010
Paluch, E., Rewak-Soroczyńska, J., Jędrusik, I., Mazurkiewicz, E., & Jermakow, K. J. A. M. (2020). Prevention of biofilm formation by quorum quenching. Applied Microbiology and Biotechnology, 104, 1871–1881.
Pauling, L., Itano, H. A., Singer, S. J., & Wells, I. C. (1949). Sickle Cell Anemia, a Molecular Disease. Science, 110(2865), 543–548. http://www.jstor.org/stable/1676635http://www.jstor.org/stable/1676635
Pavlin, J. A., Gilchrist, M. J., Osweiler, G. D., & Woollen, N. E. (2002). Diagnostic analyses of biological agent-caused syndromes: laboratory and technical assistance. Emergency Medicine Clinics of North America, 20(2), 331–350. https://doi.org/10.1016/s0733-8627(01)00004-9https://doi.org/10.1016/s0733-8627(01)00004-9
Petrosino, J. F., Highlander, S., Luna, R. A., Gibbs, R. A., & Versalovic, J. (2009). Metagenomic pyrosequencing and microbial identification. Clinical Chemistry, 55, 856–866. https://doi.org/10.1373/clinchem.2008.107565https://doi.org/10.1373/clinchem.2008.107565
Plante, J. A., Liu, Y., Liu, J., Xia, H., Johnson, B. A., Lokugamage, K. G., Zhang, X., Muruato, A. E., Zou, J., Fontes-Garfias, C. R., Mirchandani, D., Scharton, D., Bilello, J. P., Ku, Z., An, Z., Kalveram, B., Freiberg, A. N., Menachery, V. D., Xie, X., Plante, K. S., … Shi, P. Y. (2021). Spike mutation D614G alters SARS-CoV-2 fitness. Nature, 592(7852), 116–121. https://doi.org/10.1038/s41586-020-2895-3https://doi.org/10.1038/s41586-020-2895-3
Raoult, D., Fournier, P. & Drancourt, M. (2004). What does the future hold for medical microbiology?. Nature Reviews Microbiology, 2, 151–159. https://doi.org/10.1038/nrmicro820https://doi.org/10.1038/nrmicro820
Robbins, P. F., Morgan, R. A., Feldman, S. A., Yang, J. C., Sherry, R. M., Dudley, M. E., Wunderlich, J. R., Nahvi, A. V., Helman, L. J., Mackall, C. L., Kammula, U. S., Hughes, M. S., Restifo, N. P., Raffeld, M., Lee, C. C., Levy, C. L., Li, Y. F., El-Gamil, M., Schwarz, S. L., Laurencot, C., … Rosenberg, S. A. (2011). Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology, 29(7), 917–924. https://doi.org/10.1200/JCO.2010.32.2537https://doi.org/10.1200/JCO.2010.32.2537
Rogers, G. B., Stressmann, F. A., Walker, A. W., Carroll, M. P., & Bruce, K. D. (2010). Lung infections in cystic fibrosis: Deriving clinical insight from microbial complexity. Expert Review of Molecular Diagnostics, 10, 187–196. https://doi.org/10.1586/erm.09.81https://doi.org/10.1586/erm.09.81
Romano, M., Ruggiero, A., Squeglia, F., Maga, G., & Berisio, R. (2020). A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells, 9(5), 1267. https://doi.org/10.3390/cells9051267https://doi.org/10.3390/cells9051267
Rutherford, S. T., & Bassler, B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harbor Perspectives in Medicine, 2(11), a012427. https://doi.org/10.1101/cshperspect.a012427https://doi.org/10.1101/cshperspect.a012427
Sauer, B., & Henderson, N. (1988). Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proceedings of the National Academy of Sciences of the United States of America, 85(14), 5166–5170. https://doi.org/10.1073/pnas.85.14.5166https://doi.org/10.1073/pnas.85.14.5166
Savitskaia K. I. (1993). Klinicheskaia mikrobiologiia: struktura i funktsii spetsializirovannykh laboratoriĭ [Clinical microbiology: the structure and functions of specialized laboratories]. Antibiotiki i khimioterapiia = Antibiotics and chemoterapy [sic], 38(10–11), 49–55.
Saxton, R. A., Glassman, C. R., & Garcia, K. C. (2023). Emerging principles of cytokine pharmacology and therapeutics. Nature reviews. Drug Discovery, 22(1), 21–37. https://doi.org/10.1038/s41573-022-00557-6https://doi.org/10.1038/s41573-022-00557-6
Schäfer, F., Seip, N., Maertens, B., Block, H., & Kubicek, J. (2015). Purification of GST-Tagged Proteins. Methods in Enzymology, 559, 127–139. https://doi.org/10.1016/bs.mie.2014.11.005https://doi.org/10.1016/bs.mie.2014.11.005
Sharma, K., Mishra, A. K., Mehraj, V., & Duraisamy, G. S. (2014). Advances and applications of molecular cloning in clinical microbiology. Biotechnology & Genetic Engineering Reviews, 30(1–2), 65–78. https://doi.org/10.1080/02648725.2014.921501https://doi.org/10.1080/02648725.2014.921501
Sinha, R., & Shukla, P. (2019). Antimicrobial Peptides: Recent Insights on Biotechnological Interventions and Future Perspectives. Protein and Peptide Letters, 26(2), 79–87. https://doi.org/10.2174/0929866525666181026160852https://doi.org/10.2174/0929866525666181026160852
Smith, J. D. (2016). Human Macrophage Genetic Engineering. Arteriosclerosis, Thrombosis, and Vascular Biology, 36(1), 2–3. https://doi.org/10.1161/ATVBAHA.115.306796https://doi.org/10.1161/ATVBAHA.115.306796
Tay, W. H., Chong, K. K., & Kline, K. A. (2016). Polymicrobial-Host Interactions during Infection. Journal of Molecular Biology, 428(17), 3355–3371. https://doi.org/10.1016/j.jmb.2016.05.006https://doi.org/10.1016/j.jmb.2016.05.006
Telenti, A., Honore, N., Bernasconi, C. A., March, J., Ortega, A., Heym, B., … & Cole, S. T. (1997). Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. Journal of Medical Microbiology, 35(3), 719–723.
Tiller T. (2011). Single B cell antibody technologies. New Biotechnology, 28(5), 453–457. https://doi.org/10.1016/j.nbt.2011.03.014https://doi.org/10.1016/j.nbt.2011.03.014
Turner, A. K., Begon, M., Jackson, J. A., Bradley, J. E., & Paterson, S. (2011). Genetic diversity in cytokines associated with immune variation and resistance to multiple pathogens in a natural rodent population. PLoS Genetics, 7(10), e1002343. https://doi.org/10.1371/journal.pgen.1002343https://doi.org/10.1371/journal.pgen.1002343
Upchurch, K. S., & Kay, J. (2012). Evolution of treatment for rheumatoid arthritis. Rheumatology (Oxford, England), 51 Suppl 6, vi28–vi36. https://doi.org/10.1093/rheumatology/kes278https://doi.org/10.1093/rheumatology/kes278
Vadakkan, K. (2020). Molecular mechanism of bacterial quorum sensing and its inhibition by target specific approaches. In Quorum Sensing: Microbial Rules of Life (pp. 221–234). American Chemical Society.
Vadakkan, K., Choudhury, A. A., Gunasekaran, R., Hemapriya, J., & Vijayanand, S. (2018). Quorum sensing intervened bacterial signaling: pursuit of its cognizance and repression. Journal of Genetic Engineering and Biotechnology, 16(2), 239–252.
Verhoef, L. M., van den Bemt, B. J., van der Maas, A., Vriezekolk, J. E., Hulscher, M. E., van den Hoogen, F. H., Jacobs, W. C., van Herwaarden, N., & den Broeder, A. A. (2019). Down-titration and discontinuation strategies of tumour necrosis factor-blocking agents for rheumatoid arthritis in patients with low disease activity. The Cochrane Database of Systematic Reviews, 5(5), CD010455. https://doi.org/10.1002/14651858.CD010455.pub3https://doi.org/10.1002/14651858.CD010455.pub3
Wang, P., Nair, M. S., Liu, L., Iketani, S., Luo, Y., Guo, Y., Wang, M., Yu, J., Zhang, B., Kwong, P. D., Graham, B. S., Mascola, J. R., Chang, J. Y., Yin, M. T., Sobieszczyk, M., Kyratsous, C. A., Shapiro, L., Sheng, Z., Huang, Y., & Ho, D. D. (2021). Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. BioRxiv: The Preprint Server for Biology, 2021.01.25.428137. https://doi.org/10.1101/2021.01.25.428137https://doi.org/10.1101/2021.01.25.428137
Wang, R., Simoneau, C. R., Kulsuptrakul, J., Bouhaddou, M., Travisano, K. A., Hayashi, J. M., Carlson-Stevermer, J., Zengel, J. R., Richards, C. M., Fozouni, P., Oki, J., Rodriguez, L., Joehnk, B., Walcott, K., Holden, K., Sil, A., Carette, J. E., Krogan, N. J., Ott, M., & Puschnik, A. S. (2021). Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell, 184(1), 106–119.e14. https://doi.org/10.1016/j.cell.2020.12.004https://doi.org/10.1016/j.cell.2020.12.004
Wang, X. J., Wang, X. M., Teng, D., Zhang, Y., Mao, R. Y., & Wang, J. H. (2014). Recombinant production of the antimicrobial peptide NZ17074 in Pichia pastoris using SUMO3 as a fusion partner. Letters in Applied Microbiology, 59(1), 71–78. https://doi.org/10.1111/lam.12246https://doi.org/10.1111/lam.12246
Wheat, C. W. (2010). Rapidly developing functional genomics in ecological model systems via 454 transcriptome sequencing. Genetica, 138, 433–451. https://doi.org/10.1007/s10709-008-9326-yhttps://doi.org/10.1007/s10709-008-9326-y
Williams, R. C. (1989). Restriction fragment length polymorphism (RFLP). American Journal of Physical Anthropology, 32(S10), 159–184. https://doi.org/10.1002/ajpa.1330320508https://doi.org/10.1002/ajpa.1330320508
Xia, L., Zhang, F., Liu, Z., Ma, J., & Yang, J. (2013). Expression and characterization of cecropinXJ, a bioactive antimicrobial peptide from Bombyx mori (Bombycidae, Lepidoptera) in Escherichia coli. Experimental and Therapeutic Medicine, 5(6), 1745–1751. https://doi.org/10.3892/etm.2013.1056https://doi.org/10.3892/etm.2013.1056
Xiang, X., Qian, K., Zhang, Z., Lin, F., Xie, Y., Liu, Y., & Yang, Z. (2020). CRISPR-cas systems based molecular diagnostic tool for infectious diseases and emerging 2019 novel coronavirus (COVID-19) pneumonia. Journal of Drug Targeting, 28(7–8), 727–731. https://doi.org/10.1080/1061186X.2020.1769637https://doi.org/10.1080/1061186X.2020.1769637
Yan, W. X., Hunnewell, P., Alfonse, L. E., Carte, J. M., Keston-Smith, E., Sothiselvam, S., Garrity, A. J., Chong, S., Makarova, K. S., Koonin, E. V., Cheng, D. R., & Scott, D. A. (2019). Functionally diverse type V CRISPR-Cas systems. Science (New York, N.Y.), 363(6422), 88–91. https://doi.org/10.1126/science.aav7271https://doi.org/10.1126/science.aav7271
Yang, S., & Rothman, R. E. (2004). PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. The Lancet. Infectious Diseases, 4(6), 337–348. https://doi.org/10.1016/S1473-3099(04)01044-8.https://doi.org/10.1016/S1473-3099(04)01044-8
Yang, Y., Liu, J., & Zhou, X. (2021). A CRISPR-based and post-amplification coupled SARS-CoV-2 detection with a portable evanescent wave biosensor. Biosensors & Bioelectronics, 190, 113418. https://doi.org/10.1016/j.bios.2021.113418https://doi.org/10.1016/j.bios.2021.113418
Yuan, M., Liu, H., Wu, N. C., & Wilson, I. A. (2021). Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochemical and Biophysical Research Communications, 538, 192–203. https://doi.org/10.1016/j.bbrc.2020.10.012https://doi.org/10.1016/j.bbrc.2020.10.012
Zhang C. (2021). Flare-up of cytokines in rheumatoid arthritis and their role in triggering depression: Shared common function and their possible applications in treatment (Review). Biomedical Reports, 14(1), 16. https://doi.org/10.3892/br.2020.1392https://doi.org/10.3892/br.2020.1392
Zhang, J., Tarbet, E. B., Toro, H., & Tang, D. C. (2011). Adenovirus-vectored drug-vaccine duo as a potential driver for conferring mass protection against infectious diseases. Expert Review of Vaccines, 10(11), 1539–1552. https://doi.org/10.1586/erv.11.141https://doi.org/10.1586/erv.11.141
Zhang, X., Li, F., Awan, F., Jiang, H., Zeng, Z., & Lv, W. (2021). Molecular Epidemiology and Clone Transmission of Carbapenem-Resistant Acinetobacter baumannii in ICU Rooms. Frontiers in Cellular and Infection Microbiology, 11, 633817. https://doi.org/10.3389/fcimb.2021.633817https://doi.org/10.3389/fcimb.2021.633817
Zhou, D., Chan, J. F., Zhou, B., Zhou, R., Li, S., Shan, S., Liu, L., Zhang, A. J., Chen, S. J., Chan, C. C., Xu, H., Poon, V. K., Yuan, S., Li, C., Chik, K. K., Chan, C. C., Cao, J., Chan, C. Y., Kwan, K. Y., Du, Z., … Chen, Z. (2021). Robust SARS-CoV-2 infection in nasal turbinates after treatment with systemic neutralizing antibodies. Cell Host & Microbe, 29(4), 551–563.e5. https://doi.org/10.1016/j.chom.2021.02.019https://doi.org/10.1016/j.chom.2021.02.019
Zhou, D., Zhou, R., & Chen, Z. (2021). Human neutralizing antibodies for SARS-CoV-2 prevention and immunotherapy. Immunotherapy Advances, 2(1), ltab027. https://doi.org/10.1093/immadv/ltab027https://doi.org/10.1093/immadv/ltab027
Zhu, Y., Feng, F., Hu, G., Wang, Y., Yu, Y., Zhu, Y., Xu, W., Cai, X., Sun, Z., Han, W., Ye, R., Qu, D., Ding, Q., Huang, X., Chen, H., Xu, W., Xie, Y., Cai, Q., Yuan, Z., & Zhang, R. (2021). A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nature Communications, 12(1), 961. https://doi.org/10.1038/s41467-021-21213-4https://doi.org/10.1038/s41467-021-21213-4